Introductory Biology Students’ Conceptual Models and Explanations of the Origin of Variation
Abstract
Mutation is the key molecular mechanism generating phenotypic variation, which is the basis for evolution. In an introductory biology course, we used a model-based pedagogy that enabled students to integrate their understanding of genetics and evolution within multiple case studies. We used student-generated conceptual models to assess understanding of the origin of variation. By midterm, only a small percentage of students articulated complete and accurate representations of the origin of variation in their models. Targeted feedback was offered through activities requiring students to critically evaluate peers’ models. At semester's end, a substantial proportion of students significantly improved their representation of how variation arises (though one-third still did not include mutation in their models). Students’ written explanations of the origin of variation were mostly consistent with their models, although less effective than models in conveying mechanistic reasoning. This study contributes evidence that articulating the genetic origin of variation is particularly challenging for learners and may require multiple cycles of instruction, assessment, and feedback. To support meaningful learning of the origin of variation, we advocate instruction that explicitly integrates multiple scales of biological organization, assessment that promotes and reveals mechanistic and causal reasoning, and practice with explanatory models with formative feedback.
INTRODUCTION
In Chapter 5 of On the Origin of Species, Darwin wrote:
Our ignorance of the laws of variation is profound. Not in one case out of a hundred can we pretend to assign any reason why this or that part varies more or less from the same part in the parents. (Darwin, 1859, Ch. 5, p. 167)
Although Darwin observed and described a great deal of variation among and within species, his failure to explain the mechanisms underlying the origin of variation and its inheritance contributed to skepticism about his work (Charlesworth and Charlesworth, 2009). At the same time, Gregor Mendel, unaware of Darwin's work, had the keen insight that heredity may be
the one correct way of finally reaching a solution to a question whose significance for the evolutionary history of organic forms cannot be underestimated. (Mendel, quoted in Charlesworth and Charlesworth, 2009, p. 758)
As Mendel anticipated, genetics (and much later molecular biology) clarified in great detail the biological mechanisms of variation and inheritance, leading to development of the modern synthesis (Gregory, 2009). Biologists’ current understanding of why and how evolution by natural selection occurs can be deconstructed into two fundamental principles: 1) new phenotypes arise within a population by random genetic mutation; and 2) environmental factors select phenotypic variants that are best fit to their environment; these variants become more frequent in the population over time (Bishop and Anderson, 1990; Gregory, 2009). Despite its centrality to all of biology, evolution remains conceptually difficult for biology learners at all grade levels (Smith, 2010), including college (Kalinowski et al., 2010). Efforts to develop effective tools for assessing undergraduate students’ understanding of evolution have produced forced-choice instruments like the Concept Inventory of Natural Selection (CINS; Anderson et al., 2002), constructed-response questions (Bishop and Anderson, 1990; Nehm and Reilly, 2007; Nehm et al., 2012; Opfer et al., 2012), and oral interview protocols (Nehm and Schonfeld, 2008). Beyond the format differences, these instruments converge on testing students’ understanding of closely overlapping sets of five to 10 core concepts traversing multiple levels of biological organization. All these key concept sets invariably include three genetics principles: 1) the genetic origin of variation, 2) heredity, and 3) change in heritable trait frequency in a population over time.
In a previous study, we examined college introductory biology students’ constructed explanations of evolution by natural selection (Bray Speth et al., 2009). We observed that students’ explanations were largely centered on mechanisms operating at the organismal level, with little or no attention to molecular-level causes and effects. The majority of students explicitly referred to phenotypic variation among individuals as the starting point for evolution by natural selection, but very few attempted to explain why and how that variation came to exist in the first place. This finding was largely consistent with analogous conclusions reported by Nieswandt and Bellomo (2009). Even postinstruction, only a small fraction of students (19%) explicitly referred to the molecular and genetic causes of variation in their explanations (Bray Speth et al., 2009). A similar observation was reported in a study that compared novice and expert explanations of evolution (Nehm and Ridgway, 2011). While the majority of experts consistently included heredity and the genetic origin of variation in their explanations of natural selection, only 10% of undergraduate biology students in the study incorporated these concepts across multiple explanations. Kalinowski et al. (2010) explicitly described the difficulty college students encounter when required to use molecular genetic concepts to make sense of evolution. They advocated classroom practices that elicit students’ preconceptions and promote conceptual change by helping students construct explanatory frameworks that reveal explicit connections among concepts. In addition, they proposed that students should apply their explanatory frameworks iteratively to multiple contexts and that these frameworks must include both genetics and evolutionary concepts for students to make sense of the entire process from genes to populations.
Models for Promoting and Assessing Understanding of Evolution
Scientists routinely use models to organize and communicate their knowledge and to represent complex processes and systems in a simplified way. Models are abstractions or representations of natural systems that have explanatory and predictive power (Gilbert et al., 1998; Harrison and Treagust, 2000; Schwarz et al., 2009). Model-building and model-based pedagogies were shown to promote deep and meaningful learning in the science classroom (Gobert and Buckley, 2000; Schwarz and White, 2005; Brewe, 2008) and have emerged in recent years as effective approaches for teaching and learning physiology, ecology, and cell biology (Hmelo-Silver et al., 2007; Verhoeff et al., 2008).
We developed a model-based pedagogy to teach genetics, evolution, and ecology in a large-enrollment introductory biology course for life sciences majors at a research university with very high research activity (Long et al., 2014). Students in our course constructed models as paper-and-pencil artifacts visually similar to concept maps (semantic networks of concepts, represented in boxes, interconnected by arrows indicating the relationships among them). Concept maps, widely used in educational settings as effective ways of organizing and representing domain knowledge (Novak, 1998; Novak and Canas, 2008; Jonassen, 2006), are traditionally not intended as tools for modeling dynamic systems or for explaining how systems accomplish their functions.
Drawing from a theoretical framework on systems (structure–behavior–function [SBF]; Goel et al., 1996) derived from artificial intelligence, we articulated a set of conventions that supported student-generated diagrams that, in essence, are models of biological systems (Dauer et al., 2013). The SBF framework was originally designed to describe complex engineered systems (Goel et al., 1996) and was later used for facilitating systems thinking and analyzing students’ conceptual representations of biological systems and processes (Jordan et al., 2008; Liu and Hmelo-Silver, 2009; Vattam et al., 2011). According to SBF theory, a system has structures (the parts or elements of the system), behaviors (the mechanisms operating within a system), and functions (the overall roles or outputs of the system).
On several occasions throughout the course, students were required to construct SBF-based models (see Methods) representing the origin of genetic variation, the resulting phenotypic variation, and the effect of phenotypic variation on fitness in populations subject to natural selection; we referred to these as gene-to-evolution (GtE) models. Students’ GtE models changed over the course of the semester: individual propositions within models became increasingly more correct, while models grew in complexity during the first half of the semester but became more parsimonious and accurate toward the end (Dauer et al., 2013).
In this study, we focus on analyzing how students articulated the function of their GtE models, specifically how they represented variation and the origin of variation. As students’ models became more complex and individual propositions became more accurate, we hypothesized that students’ ability to convey the overall model function would also improve. We analyzed models constructed by students on their midterm and final exams to investigate 1) whether students’ GtE models represented variation and its molecular origin, 2) how accurately students incorporated the concept of mutation into their models, and 3) whether students consistently articulated the mechanism of mutation across different types of assessment (models and short answers).
METHODS
Study Context
We conducted this study in one section (n = 182) of a large-enrollment introductory biology course for life sciences majors at a research university with very high research activity (McCormick and Zhao, 2005). The course was one semester of a two-semester introductory biology sequence that students could complete in a nonprescribed order. Student attendance was 86.7 ± 10.2% across the entire semester, based on course enrollment (n = 182). Two instructors team-taught the class and participated in all class meetings. The class met three times a week for 50-min periods for 15 weeks. Course content included principles of genetics, evolution, and ecology. Evolution served as a thread throughout the course, as students learned 1) how information contained in genes is reflected in organisms’ phenotypes, 2) how phenotype determines the differential success of individuals in different environments, and ultimately, 3) how evolutionary mechanisms, including selection, lead to population change over time. The instructional strategy was based on the iterative practice of constructing, evaluating, and revising box-and-arrow SBF-based models of biological systems. Students learned early in the course to construct models of systems by representing the physical components of a system (structures) as nouns in boxes and interconnecting them with labeled arrows indicating the mechanisms or relationships (behaviors) that lead the system to produce a given output or function (Figure 1). We adopted concept-mapping terminology to refer to each box-arrow-box “sentence” as a proposition, which is the smallest meaningful unit into which models can be decomposed for analysis (Pearsall et al., 1997; Martin et al., 2000). The SBF framework for creating and interpreting conceptual models was explicitly communicated to students and was referred to frequently throughout the course. A more detailed description of how modeling was introduced to students early in the semester, and how modeling activities became increasingly more complex over time, is reported in Dauer et al. (2013). Typically, modeling problems required students to represent one or more outputs or functions of the system under study and were scaffolded by providing a minimum set of structures that needed to be included in the model (Figure 1).
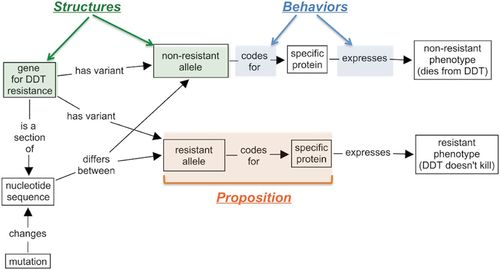
Figure 1. Example of a student-generated SBF model (transcribed in cMapTools), labeled to illustrate the model components (in italics). The example illustrates how an SBF model is a semantic network of structures (in boxes, highlighted in green) linked by behaviors (on arrows, highlighted in blue). Each box-arrow-box group (such as the one highlighted in orange) should be readable as a stand-alone unit of meaning (a proposition). This model was developed in response to a prompt asking students to represent the origin of genetic variation and resulting phenotypic variation in a mosquito population that evolved resistance to DDT. The assignment was scaffolded by providing the structures: gene, allele, nucleotides (or nucleotide sequence), protein, and phenotype.
For the purpose of this study, we analyzed GtE models that students produced in the context of the midterm and final exams. We analyzed only data from students who completed the modeling task on both exams (n = 170). The study was reviewed and determined exempt by the local IRB (protocol: IRB #X07-981).
Timeline of Instruction, Feedback, and Assessment Data Collection
Instruction on principles of evolution started in week 6, immediately following principles of genetics (Supplemental Figure S1). In week 7, instruction focused on mutation as the origin of variation. Because a detailed overview of different molecular types of mutation and of DNA repair mechanisms was beyond the scope of the course, instruction exclusively focused on point mutations as a mechanism for random changes generating new alleles. Students engaged in building and evaluating models that illustrated the origin of variation in a population of snails displaying a wide variety of shell colors and patterns. In class, students worked in groups to develop models of the origin of variation in the snail population and turned the models in to the instructors. Instructors selected four representative student-generated models for use in the following class meeting as the basis for group and whole-class discussion. Students were tasked with evaluating whether each of the four models represented the origin of variation and what possible modifications might add to or correct a model. Groups were called to report on the four models and were encouraged to propose alternative ways of representing the overall function or individual relationships. During the in-class discussion, the instructors annotated the slides to capture key points of the conversation and to underscore 1) the importance of incorporating mutation and 2) structures to which the mutation should connect. The discussion on students’ models was immediately followed by direct instruction on the molecular mechanisms of mutation. The annotated slides were made available to students after class (Supplemental Figure S2).
Midterm Exam.
The midterm exam in week 8 followed instruction, modeling practice, and feedback on the origin of variation. The entire exam was structured around the case of evolution of DDT resistance in mosquito populations (Hemingway and Ranson, 2000) and included questions on genetics and evolution and a GtE modeling problem (Table 1). Students were provided with background information on the emergence of DDT-resistant mosquito populations following widespread spraying of DDT promoted in the 1950s by the World Health Organization in an effort to eradicate malaria. To simplify the problem, we attributed DDT resistance to a single locus (R), with the recessive allele (r) causing resistance to DDT.
| Midterm exam | Final exam |
|---|---|
| Case study of DDT resistance in mosquitoes | Case study of vertebrae malformations in wolves |
| Model prompt: | Model prompt: |
| In the space below, construct a box-and-arrow model with the function: origin of genetic variation and resulting phenotypic variation in the context of this problem about DDT resistance in mosquitoes. Use language in the structures and behaviors of your model that is specific to this case. | In the space below, construct a box-and-arrow model (structures linked by behaviors) that shows the relationships among concepts that are relevant to the incidence of malformed vertebrae in wolves. |
| Include the following structures in your model: | Your model will have three functions. It will show: |
| gene, allele, protein, phenotype, nucleotides (or nucleotide sequence) | 1. The origin of genetic variation among wolves; |
| To make your model specific to this problem, you may: | 2. The relationship between genetic variation and phenotypic variation in wolves, and |
| • Use structures more than once; | 3. The consequence of phenotypic variation on wolf fitness. |
| • Add additional structures not included in the list; and, | Include the following structures in your model, but modify your language to make them specific to the case of wolves’ vertebrae. You may use structures more than once and add additional structures not in the list. |
| • Modify structures to make them specific to this case. | gene, allele, DNA, protein, phenotype, nucleotides (or nucleotide sequence), fitness |
Instruction and Feedback Following the Midterm Exam.
In the class meetings following the midterm exam (week 8), instruction on evolution continued with case studies and activities on fitness and natural selection. The midterm exam was returned in week 9, and the class received specific formative feedback on the exam GtE models. Feedback was provided in a classroom activity focused on peer evaluation of a set of four models produced by students on the exam. Student models were scanned and presented to the class as PowerPoint slides. For each model, students needed to evaluate whether and in what part of the model the origin of variation was represented (Supplemental Figure S4). Students answered each question individually with their clickers and then discussed their choices in small groups. Instructors facilitated follow-up classroom discussion to elicit the reasoning behind the consensus answers and offered clarifications when necessary.
Final Exam.
A cumulative final exam followed the unit on ecology and was structured as a series of problems based on the case of the wolves of Isle Royale, Michigan (www.isleroyalewolf.org;Räikkönen et al., 2009). Students were provided with information on an isolated population of wolves living on an island in Lake Superior. Some of the wolves have malformed vertebrae, a hereditary condition due to a change a gene (G) that regulates vertebrae formation. The allele responsible for the malformed vertebrae phenotype is recessive (g) (Bray Speth et al., 2010; Dauer et al., 2013). On the basis of this information, students were asked to construct a box-and-arrow GtE model specific to this case (Table 1). In addition, students completed a blank table by writing short answers explaining how each of five key concepts of evolution by natural selection applied to the same case. The five key concepts were: phenotypic variation, origin of variation, inheritance, fitness, and change in the population (Bray Speth et al., 2009).
| 1. The model explicitly represents variation at the genetic level (different alleles). |
| 2. The model explicitly represents variation at the phenotypic level (different phenotypes). |
| 3. Phenotypic variation is directly connected to genetic variation (e.g., there is a direct flow of information from alleles, or genotypes, to the corresponding phenotypes). |
| 4. The model includes the concept of mutation (as a structure or as a behavior). |
| a. The concept of mutation is linked to appropriate molecular-level structure/s (nucleotides, nucleotide sequence, DNA, gene, and/or allele); |
| b. Mutation is appropriately incorporated (4a above is true) and is used to explain the origin of different alleles (e.g., mutation alters a gene sequence to cause the origin of a new allele). |
Data Analysis
We developed a set of rubrics to analyze students’ models in response to three research questions:
Do Student Models Represent Variation and Its Origin?
A model function rubric (Table 2) was developed to assess whether students’ models addressed the prompt questions, meaning that they explicitly represented 1) the presence of genetic and phenotypic variation in a population and 2) the molecular mechanism leading to variation (mutation). Two raters independently applied the rubric to more than 30% of the students’ midterm models; raters had 95% interrater reliability (calculated as percent of agreement) after coding the first 30 models. Following discussion, the two raters independently coded another 30 models; the cumulative interrater agreement for all 60 models was 97%. Given the high degree of agreement, a single rater coded the remainder of the models. We calculated a total “function” score for each student model as the sum of all six items in the model function rubric (Table 2). A model that completely conveyed the required functions (represented genetic variation, phenotypic variation, and mutation as the cause of variation) would have a total score of 6.
How Accurately Do Students Connect Mutation to Other Concepts?
We analyzed the models that included mutation to evaluate how accurately students incorporated the concept into their models. We used grounded theory (Glaser and Strauss, 1967) to develop an analytical rubric. Several biologists (including the authors E.B.S., A.R., R.T., J.L.M., and T.L.) independently rated all behaviors cited by students in their GtE models (Dauer et al., 2013). The group had multiple rounds of discussion to reach a consensus over the rubric criteria. The rubric (Figure S3) assigned: 3 points to a behavior that was correct and as accurate as we would expect after instruction in an introductory biology course; 2 points to a behavior that was imprecise, poorly worded, or ambiguous but not obviously incorrect; 1 point to a biologically incorrect, unacceptable behavior or to an unlabeled arrow. For this study, we extracted, transcribed, and analyzed all the “structure-behavior-structure” propositions that included the concept of mutation as either a structure or a behavior. Two raters independently coded mutation-containing propositions for 30% of all models that incorporated mutation (n = 176 models, including 66 from the midterm exam and 110 from the final exam). Each rater scored each behavior used by students to link mutation to other biological concepts; the two raters independently assigned the same score to 92.8% of the individual propositions analyzed. Because student models had a variable number of propositions containing mutation, we calculated mean accuracy scores for each model as the sum of all points assigned to mutation-containing propositions divided by the number of propositions (e.g., Student A used mutation in two propositions, rated 2 and 3 points; her mean score was (2 + 3)/2 = 2.5). Two raters calculated mean accuracy scores for 30% of all models that incorporated mutation. Because these values were ratios, we estimated interrater reliability with a Spearman rank-order correlation coefficient test (rs = 0.85, p < 0.000001), rather than as percent of agreement. Due to the high degree of agreement, a single rater coded the remaining models.
Are Students’ Models of the Origin of Variation Consistent with Their Short Answers?
On the final exam, students completed a table with short explanations of how each of five key concepts of evolution by natural selection (phenotypic variation, origin of variation, heredity, fitness, and change in population) applied to the case of the wolves of Isle Royale. Students had previously practiced completing a similarly structured table (on an in-class quiz). We applied the “mutation” part of our function rubric (Table 2, items 4, 4a, and 4b) to code students’ short answers for the “origin of variation” concept on a 0–3 scale (0 = no mention of mutation; 1 = mutation is mentioned; 2 = mutation is mentioned and connected to genetic structures like DNA, nucleotide sequence, etc.; 3 = mutation is articulated as the causal event leading to new alleles). For example, a statement like “a random mutation sometime in the population” would receive 1 point; “a mutation in the nucleotide sequence” would receive 2 points; “The origin of variation came from a random mutation in the nucleotide sequence which resulted in the g allele” would receive 3 points. Interrater reliability was established as 95.6% (percent agreement) between two raters for more than 30% of student answers, and a single rater coded the remaining answers.
RESULTS
Do Student Models Represent Variation and Its Origin?
We analyzed students’ midterm GtE models to determine what proportion of our students explicitly represented alternative alleles (genetic variation) and phenotypes (phenotypic variation) in a population of mosquitoes undergoing natural selection for DDT resistance. Sixty-nine percent of students represented genetic variation and 60% represented phenotypic variation, but only 53% of students’ midterm models illustrated a direct flow of reasoning (possibly including “protein” as intermediate structure) from the specific allele/genotype to the corresponding phenotype. At midterm, most students failed to represent mutation as the mechanism causing variation: only 39% of all models included the concept of mutation, and an even smaller subset of these clearly represented mutation as the origin of variation (Table 3).
| Model included: | Midterm exam | Final exam |
|---|---|---|
| 1. Genetic variation (different alleles) | 69% | 76% |
| 2. Phenotypic variation (different phenotypes) | 60% | 68% |
| 3. Direct genotype–phenotype connection | 53% | 55% |
| 4. Mutation: | 39% | 65%* |
| a. linked to genetic concepts | 17% | 26% |
| b. linked to genetic concepts and used to explain the origin of new alleles | 20% | 35%* |
The proportion of students who explicitly represented genetic variation, phenotypic variation, and a direct genotype-to-phenotype connection increased on the final exam, although not significantly. The number of student models incorporating mutation, however, increased significantly on the final compared with the midterm (from 39 to 65%; chi-square test of independence, p < 0.0001). Concurrently, we observed a significant increase in the proportion of models conveying that mutation was the causal event directly responsible for a new allele in the population (from 20 to 35%; chi-square test of independence, p = 0.0024).
The mean function score for the class was 2.78 ± 1.85 (SD) at midterm and increased to 3.62 ± 1.92 (SD) on the final. For ease of representation, we grouped models that had very low function scores (0–1 points), average models (2–3 points), above-average models (4–5 points), and excellent models (6 points). From the midterm to the final exam, we observed a decrease in models scoring 0–1 and 2–3 points and an increase in models scoring 4–5 and 6 points (Figure 2). Most students remained in the same competency group or advanced to the next group. Several students (23.5%), however, jumped up two levels or more, while a smaller subset (14%) regressed to a lower score. As expected, the overall improvement in students’ model function from the midterm to the final exam was significant (Wilcoxon signed-rank test, n = 170, p < 0.0001).
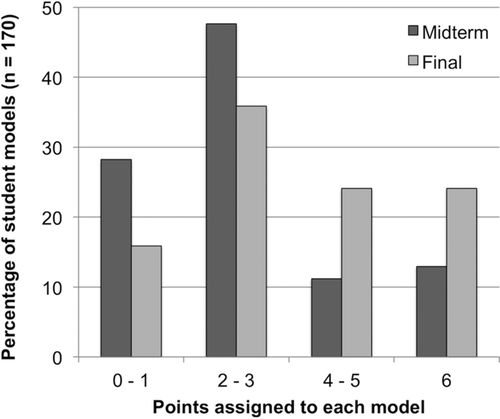
Figure 2. Change over time in student models’ function. The proportion of higher-scoring models increased on the final exam.
How Accurately Do Students Connect Mutation to Other Concepts?
During the course, students received explicit feedback on the importance of including mutation in their models to explain origin of variation but were not specifically instructed on whether to incorporate mutation as a structure or as a behavior. When students included the concept of mutation, they determined how to represent it. Mutation, as a mechanism, would be most appropriately represented as a behavior (on an arrow); however, students were never instructed to incorporate mutation as a behavior nor were they penalized for choosing to represent mutation as a structure (a physical component of the system). We recorded students’ choice to use mutation as a structure or as a behavior. We identified only a very small number of instances in which mutation was represented neither as a structure nor as a behavior; rather, it was used as an adjective to qualify a structure (e.g., “mutated gene” or “mutant allele”), or it was left “floating,” meaning that the student placed it in the model but did not connect it to any other concept. Parallel to the increase in overall use of the mutation concept from the midterm to the final, we observed a decrease in the percentage of models representing mutation as a structure and an increase in the percentage of models representing mutation as a behavior (Table 4). This shift aligns with the finding that accuracy of individual propositions in students’ models grew throughout the semester (Dauer et al., 2013) and contributes an additional dimension by which we can characterize this improvement.
| Mutation | Midterm exam | Final exam |
|---|---|---|
| represented as: | (n = 66) | (n = 110)a |
| Structure | 47 (71.2%) | 65 (59.1%) |
| Behavior | 19 (28.8%) | 43 (39.1%) |
In our analyses of students’ models, we were particularly interested in characterizing the propositional accuracy of the connections between mutation and genetic structures (such as allele, gene, nucleotides, DNA, or nucleotide sequence). Each model earned a single score, calculated as the mean score of all propositions including mutation (see Methods and Figure S2). To compare students’ performances on the midterm and final exam models, we binned students into four groups (Table 5). Students in group 1 (30.6% of the class) incorporated mutation into their models on both the midterm and the final exam; group 2 students (34.1% of the class) represented mutation only on the final exam model, but not on the midterm. A smaller percentage of students (group 3, 8.2%) used the concept of mutation only on the midterm, but not on the final. Finally, 27.1% of all students (group 4) did not incorporate mutation into their models on either exam.
| Groupa | n | Percent of 170 | Midterm (mean ± SD) | Final (mean ± SD) |
|---|---|---|---|---|
| 1 | 52 | 30.6 | 2.46 ± 0.64 | 2.34 ± 0.66 |
| 2 | 58 | 34.1 | n/a | 2.10 ± 0.69 |
| 3 | 14 | 8.2 | 2.08 ± 0.73 | n/a |
| 4 | 46 | 27.1 | n/a | n/a |
Group 1 students maintained consistent propositional accuracy between the two tests, with no statistical difference between their midterm and final scores (mean midterm score = 2.42; mean final score = 2.31; Wilcoxon signed-rank test, p = 0.48). Group 2 and 3 students only incorporated mutation into one of the two tests (the final or the midterm, respectively), and both groups did so with a lower propositional accuracy than their group 1 peers. Specifically, the mean accuracy score achieved by group 2 students on the final exam model was lower than that of group 1 students on the same exam (Mann-Whitney test, p = 0.06). The few students who incorporated mutation on the midterm but not on the final exam (group 3) also had a mean score that was lower than that of their group 1 peers on the same exam (Mann-Whitney test, p = 0.06).
Is the Representation of the Origin of Variation Consistent across Student Models and Short Answers?
On the final exam, students filled out a blank table listing five fundamental evolutionary concepts, including the origin of variation. Overall, 58% of students consistently incorporated the mutation concept across the two assessments, while 16% of all students lacked it in both (Figure 3A). Interestingly, nearly 19% of students included the concept of mutation in their short answers but not in their models, and 7% included mutation in the model but not in the short answer.
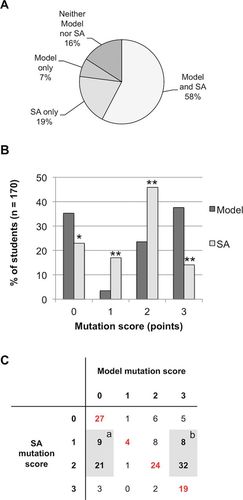
Figure 3. Use of mutation in students’ models and short answers on the final exam. (A) Consistency in the use of the mutation concept across two different assessments of the origin of variation, a model and a short-answer (SA) explanation. (B) Distribution of scores attributed to student models and short answers (SA) for their use of the mutation concept. The same rubric was applied to both assessments (Table 1, items 4, 4a, and 4b). (C) A cross-tabulation illustrating how individual student's scores were distributed in the class. Each cell indicates the number of students who had a given combination of scores on their two assessments; individuals on the red diagonal performed consistently across the two assessment questions.
We coded the short answers on a 0–3 scale, applying the same rubric we had used to code the models for the concept of mutation (see Methods and Table 1, items 4, 4a, and 4b). Using the same rubric allowed us to directly compare aggregate and individual students’ scores across two distinct assessments of the same concept (Figure 3, B and C). Aggregate score analysis (Figure 3B) revealed that the concept of mutation appeared significantly more frequently in students’ short answers than in their models (Fisher's exact test, p = 0.017). However, students incorporated mutation in short answers at a basic level (1 or 2 points) significantly more often than they did in models. Conversely, mutation was correctly incorporated as the source of new alleles in the population (3 points) significantly more frequently in students’ models than in short answers (Fisher's exact test, p < 0.0001).
Analysis of individual students’ scores on models and short answers (Figure 3C) indicated their scores on the two assessments were significantly correlated (Spearman's rs = 0.439, p = 0.01). Overall, 45% of the students had the same score on both assessments (these students are represented in red on the diagonal of the table in Figure 3C). The two largest groups that strayed from the diagonal were students who used the term “mutation” or “genetic mutation” in their short answer at a basic level (meaning they had a score of 1 or 2 on their short answers) but either failed to incorporate mutation into their model (17.6%; highlighted by the shaded box marked “a” in Figure 3C) or represented mutation in their model as the causal mechanism leading to new alleles, earning a score of 3 (23.5%; highlighted by the shaded box marked “b” in Figure 3C)
DISCUSSION
This study adds to the body of evidence that college introductory biology students struggle to integrate molecular genetic concepts within their evolutionary reasoning. Specifically, we uncovered students’ difficulty incorporating the molecular basis of variation in their explanatory frameworks of evolution by natural selection.
The literature on evolution teaching and learning is rich with evidence that evolution as a whole is conceptually difficult for students (Bishop and Anderson, 1990; Anderson et al., 2002). A recent metastudy of introductory biology students’ learning of natural selection across multiple courses and institutions reported that students achieved only modest learning gains (Andrews et al., 2011), measured by an abbreviated version of the CINS (Anderson et al., 2002) and a short constructed response (Bishop and Anderson, 1990; Nehm and Reilly, 2007).
The results of our study align with previous reports showing that students’ explanations of natural selection largely fail to incorporate molecular genetic concepts like genetic variation and heredity (Nehm and Schonfeld, 2008; Nehm and Ridgway, 2011; Bray Speth et al., 2009; Nieswandt and Bellomo, 2009). We further extend these findings with additional evidence that college introductory biology students consistently struggle to integrate the molecular basis of variation in their explanatory frameworks of evolution by natural selection. After a semester-long introductory biology course on genetics, evolution, and ecology, and despite instruction that emphasized the mechanisms underlying variation and included formative assessment and targeted feedback, nearly one-third of our students still did not incorporate mutation into their models.
Students Struggle to Represent the Origin of Variation
In week 7 of class, before the midterm exam, instruction focused extensively on mutation as the causal mechanism that generates variation. Despite modeling practice in class and explicit feedback, only 39% of all students included the concept of mutation in their midterm exam models (Table 3). Moreover, only 20% incorporated mutation as the causal mechanism explaining the origin of new alleles (Table 3). Instructors immediately identified this gap in students’ midterm models and designed a second targeted round of feedback following the midterm exam (see Methods and Figure S3). The proportion of students who incorporated mutation into their models of the origin of variation increased significantly (to 65%) on the final exam.
Mutation is an inherently difficult concept for various reasons. To begin with, it is a molecular-scale mechanism that explains organism- and population-scale outcomes. The science education literature has shown that constructing causal explanations of biological phenomena is difficult for learners. Students often resort to teleological and anthropomorphic explanations or fail to recognize the need to include causal or mechanistic reasoning when asked to articulate an explanation of biological change, particularly in the context of adaptation and evolution (Abrams and Southerland, 2001; Southerland et al., 2001; Russ et al., 2008). Studies on learning about systems have shed further light on students’ apparent difficulty with reasoning about underlying mechanisms, as these studies demonstrate that novice learners tend to focus on the perceptually salient, structural aspects of systems (Hmelo et al., 2000; Hmelo-Silver and Pfeffer, 2004), rather than their functions and behaviors. Micro-level components and implicit mechanisms pose a substantial learning challenge, especially when learners must infer them (Chi et al., 1994; Hmelo-Silver et al., 2007) and connect causal processes across multiple levels. An additional issue further complicating the understanding of mutation is that it is a random event; students often hold deep misconceptions about the role of random processes in the natural world (Garvin-Doxas and Klymkowsky, 2008). On the basis of this understanding, we argue that articulating the role of random mutation as the underlying source of variation, unless explicit cues are provided or elicited, is an inference that requires retrospection: students need to recognize that the observable, heritable phenotypic variation within a population is caused by the existence of multiple alleles and that mutation events must have occurred at the molecular level in the past, causing the new alleles to exist. A systems-thinking skills hierarchy developed in the context of learning about natural systems places retrospection (with prediction) at the top, as one of the most advanced cognitive characteristics of systems thinking (Ben-Zvi Assaraf and Orion, 2005).
It is possible that the improvement we observed in students’ ability to incorporate mutation into their models on the final exam was due, at least in part, to their developing systems-thinking skills and ability to reason causally and mechanistically about evolution. Of course, we cannot exclude that students were simply repeating information they obtained during feedback. Our data do not allow discriminating between these alternative explanations, nor do they allow us to separate students’ gains in conceptual understanding from their possible increased familiarity and proficiency with model building. However, it is evident that a single cycle of modeling, instruction, and feedback was not sufficient for the majority of the class, and after two cycles, we still observed that 35% of our students did not include mutation in their explanatory frameworks. It is noteworthy that in other reported assessments of students’ understanding of natural selection (Nehm and Reilly, 2007; Nehm and Schonfeld, 2008; Bray Speth et al., 2009), students were not explicitly prompted to incorporate mutation, and we have no evidence of whether they had received feedback on how to include this concept in their explanations. In the course described in this study, instructors repeatedly emphasized the importance of representing mutation as the source of variation; yet after repeated opportunities for practice and feedback, only 65% of students in the course incorporated mutation into their final exam models, and only 35% appropriately used it to explain the origin of new alleles (Table 3). We recognize that not all strategies for providing feedback are equally effective (Hattie and Timperley, 2007) and that it may be necessary to critically evaluate the efficacy of different mechanisms of feedback that help students incorporate this concept into their explanatory frameworks.
Student Models of the Origin of Variation Become More Meaningful over Time
Students’ final models, overall, better conveyed the function that was required in the prompt (representing variation in a population and the origin of this variation; Figure 2). Previous comprehensive analysis of all propositions in students’ models had revealed that the biological accuracy of individual propositions within models increased throughout the semester (Dauer et al., 2013). Our results support the hypothesis that, as students’ language in defining individual relationships among structures became more accurate, their ability to represent the overall system function also improved.
These results were not uniform across student groups. For students who incorporated mutation into both models (group 1), the accuracy of propositions containing the mutation concept did not significantly change between the midterm and the final exam (Table 5). We observed, however, that students who did not incorporate mutation on the midterm but only on the final exam (group 2) used less accurate propositions than their group 1 peers. While this difference did not appear to be statistically significant, the p value was close enough to significance level (p = 0.06, α = 0.05) to warrant discussion of this outcome. It is possible that students who added mutation to their models late in the semester were still tentative on how to incorporate it. We may interpret this difference in terms of stages of cognitive structure development, which proceeds by accretion—addition of new concepts to existing knowledge—followed by restructuring and tuning, major rearrangements and minor refinements of the network of relationships among new and old concepts (Vosniadou and Brewer, 1987; Pearsall et al., 1997; Dauer et al., 2013). At midterm, group 1 students had already added mutation to their cognitive structure and accommodated it within the network of relationships among concepts in their evolution reasoning framework. Group 2 students added mutation later, and at the time of the final exam, they were still possibly tuning or restructuring their conceptual models to accommodate the new concept; their propositions, thus, were less accurate than those of group 1.
In this study, we compared students’ GtE models of two different systems with clearly distinct surface features. At midterm, students modeled evolution of DDT resistance in mosquito populations (an instance of trait gain); on the final exam, the model context was that of a deleterious mutation in wolves, causing loss of ability to effectively walk and hunt (an instance of trait loss), which persisted due to isolation and inbreeding. Surface item features have been shown to affect the frequency of naïve and key concepts of evolution in students’ constructed explanations (Nehm and Ha, 2011). Generally, students tend to include fewer key concepts and more naïve conceptions in cases of trait loss than in cases of trait gain, although the differences are less pronounced in within-species contexts. On the basis of this evidence, we would have predicted that students may incorporate mutation with similar or lower frequency in their final (wolf, trait loss) than in their midterm (mosquito, trait gain) model. The higher frequency that we observed on the final exam's models, therefore, may be attributed to learning. It is possible, however, that the frequency of the mutation concept may have been even higher on the final exam, had a trait-gain problem been presented. Future studies with a split-plot design in which distinct surface features are tested simultaneously may provide further insight.
Course instructors did not specifically address whether mutation should be represented as a structure or as a behavior and did not evaluate students’ models differently based on this choice. We observed, on the final exam model, a shift toward using mutation as a behavior as opposed to a structure (Table 4). One possible explanation for this shift is that the student models shown as examples in the postmidterm feedback activity happened to have mutation placed on arrows (Figure S3). Students may have interpreted that as a suggestion for improvement. Alternatively, we could interpret students’ later preference for placing mutation on arrows as an indication of a better appropriation of the concept and of biological language. Mutation is, in fact, a mechanism, and as such, it is more appropriately represented as a behavior (not as a physical structure) of the system. Although it is common among practicing biologists to refer to an altered genetic sequence as a mutation, in the context of introductory biology, we did not explore with students the nuances of the term in its various applications. Moreover, we observed that when using “mutation” as a structure, students typically would construct propositions like “mutation changes the nucleotide sequence” or “mutation creates a new allele,” wherein mutation was represented as an abstract agent causing a change. In this context, we interpreted students’ shift toward placing mutation on arrows rather than in boxes as an example of their progress toward a more accurate understanding.
Mechanistic Reasoning about Mutation Emerges in Models More Than in Short Answers
A segment of the class population was consistent in the quality of their reasoning about mutation across their models and short answers (Figure 3). The vast majority of students who mentioned mutation in their short answers but not in their models (Figure 3C) did so at a basic level. This suggests that their understanding of mutation still may have been weak and poorly integrated within their knowledge structure. Students were able to use the word “mutation” in their short answers without further qualifying the concept or explaining how mutation led to variation, but could not do the same in models. Incorporating a concept in a model, in fact, requires building at a minimum one meaningful connection to another concept.
At the other end of the spectrum, we observed a group of students who incorporated mutation at the best possible level in their models but failed to meet the same high explanatory standard in their short answers. Again, this suggests that the format of the modeling task is more conducive for eliciting causal and mechanistic reasoning than a short answer, even in the presence of a solid understanding. Nieswandt and Bellomo (2009) came to similar conclusions: students’ written responses about evolution had a fairly low explanatory power and failed to display “schematic knowledge” (the why of a system).
Our finding that students’ written responses tend to be less conducive to mechanistic reasoning than models may represent a limitation of this study, since we did not coach students to write explanations. In an instructional context in which explanations as a form of assessment are appropriately scaffolded (McNeill et al., 2006), we might expect students to better articulate causal and mechanistic reasoning.
Implications for Teaching and Learning
Despite numerous calls for integrating evolutionary reasoning across the curriculum (American Association for the Advancement of Science [AAAS], 2011; Olson and Labov, 2012), evolution teaching and learning remains largely fragmented. Traditional textbooks and curricula present evolution as a discrete topic and provide few opportunities for students to practice making the conceptual connections across levels of biological organization necessary for a complete and accurate understanding of evolution by natural selection (Nehm et al., 2009). Instructional strategies that focus primarily on memorizing content while following textbook-driven compartmentalization of concepts do not promote the kinds of reasoning necessary to make sense of complex biological problems (National Research Council, 2003).
Learning about biology from a systems perspective is increasingly recognized as both a challenge and a priority (AAAS, 2011). In this course, we implemented a model-based pedagogy grounded in the cognitive sciences and aimed at fostering integrative and systems thinking. SBF models proved a suitable system representation tool, because this syntax overcame several limitations of concept maps (Tripto et al., 2013). The modeling approach we described in this and other studies (Dauer et al., 2013; Long et al., 2014) promotes integrative thinking, as it requires students to repeatedly articulate the connections between genetics and evolution in a number of different contexts. Additional strategies that promote integration of genetics and evolution are grounded in using authentic DNA sequences (Kalinowski et al., 2010) and case studies (White et al., 2013) incorporating well-characterized genetic mutations, the resulting phenotypes, and known evolutionary outcomes.
Along with instruction that promotes integrative thinking, assessment needs to both elicit student reasoning across levels of organization and serve as a source for frequent formative feedback in support of meaningful learning and progressive restructuring and tuning of students’ knowledge frameworks. Using models as an assessment tool allowed instructors to rapidly gauge students’ understanding of mechanisms and functions and to provide timely and targeted feedback. Student models illuminated aspects of their thinking we might have otherwise missed had we exclusively relied on other types of constructed-response assessments such as written explanations.
In summary, we advocate evolution instruction that 1) explicitly connects molecular-level processes to organism- and population-level events in the context of multiple gene-to-evolution cases; 2) relies on modes of assessment, such as conceptual modeling, that promote and reveal student reasoning about the causes and mechanisms underlying evolution by natural selection; and 3) iteratively provides opportunities for students to practice constructing and using their explanatory frameworks and to receive formative feedback on their thinking.
ACKNOWLEDGMENTS
We are very thankful to several collaborators. Diane Ebert-May and Sara Wyse collaborated in developing instruction and assessments for the course. Joe Dauer and Sara Wyse participated in group discussion and refinement of the grounded rubric, including the portion we applied in this study. We thank Laurie Russell, two anonymous reviewers, and members of the Bray Speth lab for critically reviewing the manuscript. This article is based on work supported in part by the National Science Foundation (NSF) under grants DUE-0736928 and DRL-0910278 (T.L., principal investigator). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF.



