The Python Project: A Unique Model for Extending Research Opportunities to Undergraduate Students
Abstract
Undergraduate science education curricula are traditionally composed of didactic instruction with a small number of laboratory courses that provide introductory training in research techniques. Research on learning methodologies suggests this model is relatively ineffective, whereas participation in independent research projects promotes enhanced knowledge acquisition and improves retention of students in science. However, availability of faculty mentors and limited departmental budgets prevent the majority of students from participating in research. A need therefore exists for this important component in undergraduate education in both small and large university settings. A course was designed to provide students with the opportunity to engage in a research project in a classroom setting. Importantly, the course collaborates with a sponsor's laboratory, producing a symbiotic relationship between the classroom and the laboratory and an evolving course curriculum. Students conduct a novel gene expression study, with their collective data being relevant to the ongoing research project in the sponsor's lab. The success of this course was assessed based on the quality of the data produced by the students, student perception data, student learning gains, and on whether the course promoted interest in and preparation for careers in science. In this paper, we describe the strategies and outcomes of this course, which represents a model for efficiently providing research opportunities to undergraduates.
INTRODUCTION
The Howard Hughes Medical Institute (HHMI) and others have challenged administrators of undergraduate science programs to provide more opportunities for research experiences (National Science Foundation, 2000; Wenzel and Karukstis, 2004; Anderson et al., 2011), echoing a number of articles on science education published over the past century (Mariken et al., 2009). Today, science faculties at universities in the United States name opportunities for “oral communication, design of some aspect of a project and having a meaningful research question” as essential features of an undergraduate research program, a view that is also held internationally (Lopatto, 2003).
Students who engage in novel scientific research as part of their undergraduate education demonstrate improved understanding of concepts and are more likely to exhibit a continued interest in science (Russell et al., 2007; Lopatto, 2009; Mervis, 2010). However, in practice, support for these opportunities is limited by the availability of funding and by demands on research personnel who could serve as mentors (Lopatto, 2003). Means of providing research opportunities to undergraduates include traditional summer research programs and independent study and honors thesis projects, which require individual mentorship in a faculty laboratory and, by extension, limit the number of students who can participate (Huggins et al., 2007). Creative extensions of these opportunities on a larger scale have been instituted (reviewed in Wei and Woodin, 2011) and include programs such as the Small World Initiative (Yale University Center for Scientific Teaching; Barral et al., 2014), Phage Hunters Advancing Genomics and Evolutionary Science (Jordan et al., 2014, and the Genomics Education Partnership (Shaffer et al., 2010), which are crowdsourcing projects that collectively involve more than 100 universities and thousands of undergraduates worldwide. These programs provide the basis for other models that could offer research experience to undergraduates while also creating a sense of membership in the scientific community on a global level.
The difficulties in extending research opportunities to students are particularly evident at larger universities (Wood and Gentile, 2003). For example, at the University of Colorado at Boulder, ∼120 undergraduates are receiving degrees in molecular, cellular, and developmental biology (MCDB) per year. A department of 37 research-intensive faculty (University of Colorado at Boulder, 2010) makes it impractical to offer all students majoring in MCDB research opportunities, thus reinforcing the dichotomy between undergraduate and research laboratory settings.
To provide students with an authentic laboratory research experience, we created an undergraduate laboratory-intensive course with the support from the HHMI Professors Program, the Python Project, taught by postdoctoral fellows in a structured classroom environment. Importantly, a goal of the course is to guide students through novel experimentation in conjunction with an ongoing research project in the sponsor's laboratory. The course offers students the opportunity to participate in all aspects of experimental design and data acquisition, analysis, and interpretation, thus creating an auxiliary laboratory that operates in collaboration with the sponsor's laboratory. Students also review primary literature; participate in ongoing experimental optimization; and, ultimately, produce a written report in the format of a scientific article, give an oral presentation, and present their work in a public poster session. By design, the data produced in both the class and sponsor's laboratory shape the aims of the project each semester, such that students in each semester examine expression of a unique set of genes related to organ hypertrophy or regression in the Burmese python. Thus, the course mirrors opportunities normally provided only to members of a laboratory research group and ensures that students experience the excitement and challenges of scientific discovery with the benefit of supervision and mentoring. In addition to serving the undergraduate population, this model also addresses a growing need for postdoctoral fellow involvement in science curriculum reform (Wieman, 2009).
COURSE DESIGN AND EVALUATION
Student Demographics
This report includes data obtained from undergraduate students enrolled in each of 10 semesters from 2009 to 2013 (Table 1). The maximum enrollment for each semester was 16 students, with a 1:8 instructor-to-student ratio. Current required prerequisites are introductory courses in cell biology, molecular biology, genetics, and introductory biochemistry. Instructor permission, which is obtained through a short interview or detailed email exchange, is intended to ensure that students understand the unconventional nature of the course and is required for course enrollment. Undergraduate students majoring in MCDB in their junior or senior year are eligible to enroll in the Python Project as an elective course; however, a small number of students with other majors have also been approved to register. More than half of the students were in their fourth or fifth year of undergraduate education.
| Percentage of students (n = 84) | |
|---|---|
| Male | 53.6 |
| Female | 46.4 |
| Ethnicity | |
| Underrepresented minoritiesa | 6.0 |
| Foreign exchange | 9.5 |
| Year | |
| Freshman | 7.1 |
| Sophomore | 3.6 |
| Junior | 29.8 |
| Senior | 40.5 |
| Fifth-year senior | 19.0 |
| Major | |
| MCDB | 79.8 |
| Biochemistry | 8.3 |
| Other | 11.9 |
Course Organization
Before laboratory experiments begin, basic biosafety training is provided by the instructors using University of Colorado Environmental Health and Safety (EH&S) guidelines including explanation of material safety data sheets, use of personal protective equipment (gloves, masks, eye protection), appropriate disposal of hazardous chemicals (primary and secondary containers and disposal services are provided by University of Colorado EH&S), use of UV light to visualize DNA, and general maintenance of equipment and solutions necessary to maintain a safe and orderly environment.
Students in the Python Project perform experiments in parallel with a larger project in the sponsor's laboratory that examines the molecular mechanisms regulating organ hypertrophy in Burmese pythons after feeding (Riquelme et al., 2011). To manage the systemic stress associated with digesting meals that can reach up to half of their body weight, pythons undergo several extraordinary physiological changes: increased oxygen consumption, an extreme decrease in the pH of the stomach, and significant hypertrophy of many organs (Secor, 2003, 2008). For example, the mass of the ventricle of a python heart can increase up to 40% in as little as 48 h after feeding relative to that of a fasting python, a change that is accompanied by a 40-fold increase in metabolic rate (Secor and Diamond, 1998). Rapid growth of the python organs after feeding provided a unique opportunity to examine the mechanisms responsible for organ growth in terms of expression of genes related to proliferation or hypertrophy.
Although students do not handle, care for, or dissect the Burmese pythons, they participate in harvesting organs by preparing tubes and freezing samples during a dissection performed by a postdoctoral fellow. To ensure that students understand the responsibilities associated with working with animals, instructors explain the process of gaining approval for experimentation through the Institutional Animal Care and Use Committee review, the justification for animal use, and the training required to work with animals at the University of Colorado at Boulder.
The course is organized into six laboratory hours and, on average, one 50-min lecture per week (Table 2). Currently, the first month of the semester mainly consists of instruction and supervised technical practicums. In the second and third months of the course, students work independently to design DNA primers for quantitative polymerase chain reaction (qPCR); isolate RNA, redesign primers, if necessary; and synthesize cDNA (Riquelme et al., 2011). Learning objectives are centered on the design and successful completion of a novel research project using Burmese python tissues (Table 3). Evaluation of student progress is achieved through written assessments (worksheets, midterm exam, review papers) and oral presentations (a 10-min talk introducing the background, hypothesis, and expected outcomes of their project, and a public poster session).
| Lecture 1 | Course introduction, general chemistry review |
| Lecture 2 | Sponsor's lab member lecture 1: Overview of the python project |
| Lectures 3–6 | Primer design |
| Lecture 7 | Sponsor's lab member lecture 2: Gene candidate review |
| Lecture 8 | RNA isolation and spectrophotometry |
| Lecture 9 | cDNA synthesis |
| Lecture 10 | 10-min talk: review and demonstration |
| Lecture 11 | Cloning and enzyme restriction analysis |
| Lecture 12 | Pipetting accuracy and exercise review/demonstration |
| Lectures 13–15 | Introduction to qPCR |
| Lecture 16 | Biostatistics |
| Lecture 17 | Poster examples and demonstration of presentation |
| Learning objective | Assessment |
|---|---|
| 1. Design experiments to measure gene expression in postprandial Burmese python tissues | Midterm examination, oral presentation, poster session, and research paper |
| 2. Perform experiments to validate all aspects of experimental design | In silico validation of primers in a related species, PCR validation of product size, and qPCR validation of single-product amplification |
| 3. Research background on the gene of choice using primary literature and develop a well-supported hypothesis | Review paper, oral presentation, poster session, and research paper |
| 4. Understand and be able to describe the rationale for each experiment and choice of gene | Poster session and research paper |
| 5. Develop proficiency in qPCR | qPCR experiments |
| 6. Present data effectively in a public poster presentation | Poster session |
An important aspect of the Python Project is its evolution as new resources became available. When the course began, no sequence information was available for the Burmese python. Degenerate PCR techniques were used to elucidate the transcripts of the students’ genes of interest. As more information about the python genome became available, students began to heavily rely on the transcripts of other organisms to assemble the transcripts of their genes of interest and to design primers. For verification that the primers they designed amplify the correct target sequence, primers are tested in silico using online databases and conventional PCR to assess product size (Figure 1). Other approaches include cloning and sequencing and use of restriction enzymes to validate the PCR identity of the product. In the final weeks of the semester, students’ projects diverge greatly, such that each student progresses independently, based on the success of his or her preparatory experiments. Accordingly, the first hour of laboratory periods shifts at this time to accommodate a laboratory meeting format in which students present results and receive feedback on experiments.
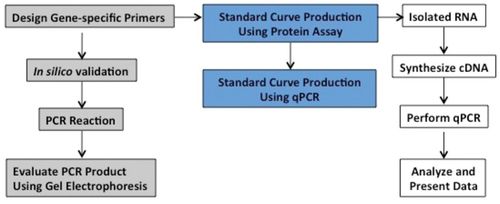
Figure 1. Flowchart of experimental design. The course requires three major processes: primer design (gray boxes), validation of appropriate techniques (blue boxes), and measurement of gene expression (white boxes). Primers are designed using the assembled transcript for the gene of interest. In silico validation includes translation of the assembled transcript and verification of the intended product amplified by the primers. Primers are also tested in vitro using conventional PCR; the students evaluate whether a product of the correct size is amplified by using their primers. While primers are being synthesized, students begin pipetting exercises to demonstrate accurate pipetting skills before beginning qPCR. These exercises include production of standard curves using a protein assay and a highly expressed control gene using qPCR. The semester culminates with qPCR experiments to measure expression of students’ genes of interest, which requires both RNA isolation and cDNA synthesis.
Students can find this type of curriculum to be challenging, because they are not accustomed to the level of independence inherent in this approach. For example, students frequently require reassurance that experimental failure does not indicate course failure, likely a product of their past laboratory experiences, in which grading was based on successful technical execution of protocols to yield known answers. Although adjusting to independence takes time, students ultimately demonstrate a high level of investment in their projects, particularly at the end of the semester, as they work toward a final result. In fact, in the most recent offering of this course, students requested 30 additional laboratory hours in the last 2 wk of the semester to complete extra experiments related to their projects.
Course Resources
All supplies required to perform experiments were provided to students (Table 4). According to University of Colorado at Boulder policy, students taking courses in the MCDB Department are required to pay a standard laboratory fee ($31); the course was initially subsidized by a grant from the HHMI (to L.A.L., HHMI Professors Program) and currently receives supplemental funding from a gift from a pharmaceutical company (Amgen). An average of $692 per student was spent for the first five semesters, which included the purchase of equipment to start the Python Project. Currently, the cost per student for disposables is approximately $172.
| Common use laboratory equipment | Bench supplies for each student |
|---|---|
| Bio-Rad CFX96 Real-Time PCR Detection System410-g top-loading balanceUltraCam 8gD Digital Imaging SystemVariable intensity, 20 cm × 20 cm, 300-nm UV transilluminatorPC with Windows XP Pro (3)Biophotometer with printerHeating blockVortex, variable speed, 120 V, 6 Hz (8)Thermal printer, 100–240 VSpectrafuge 16 M, 120 VDNA Gel SystemPipetman: P2 (4) | Tips: P10, P20, P200, P1000Pipetman: P20, P200, P100015-ml conical tube racks50-ml conical tube rackIce bucketPCR tube rack1.5-ml tube rack1.5-ml RNase-free tubesTimerIndelible markerElectrophoresis apparatusRNase-free waterSolid-waste containerLiquid-waste containerLabeling tapeClear tapeScissors |
A course manual with primary literature articles on Burmese python physiology; basic laboratory practices, protocols, and rationale for experiments (see the Supplemental Material); and guidelines for oral presentations, laboratory notebook maintenance, writing a literature review, preparing a research report, and poster preparation and presentation was provided to students without charge. Currently, these resources are offered electronically through an online learning platform, Desire2Learn. Lecture presentations, study guides, and answer keys are also available through Desire2Learn. Communication with students outside classroom hours is achieved through email; the course website; and a Python Project Facebook page that conveys lay information about the Python Project, links to scientific articles related to the research, and photographs of poster sessions from previous semesters. Students are also provided with access to the laboratory during the day when instructors are available, if extra time is requested.
Technical Skills Training
Students were monitored consistently throughout the semester through informal assessments to ensure that the appropriate technical skills were mastered before they moved on to more complex tasks. The goal was to emphasize the importance of this type of quality control before data are produced. For example, pipetting skills were tested before beginning qPCR, a technique that requires precise measurement of small volumes (4–6 μl). Through a series of exercises that required students to measure protein concentrations in a dilution series, the students produced a standard curve and determined the concentration of protein in three unknown samples. This proved to be the largest obstacle for the students, who were required to achieve a minimum R-squared value of 0.98, indicating the production of accurate dilutions and triplicate samples of each dilution. On average, students performed six titration experiments to demonstrate adequate pipetting skills, with this task spanning two to three laboratory periods. After students demonstrated accurate pipetting, we then examined their qPCR results for accuracy of technical replicates; existence of a single melt curve for their primers, indicating that the primers designed yield a single PCR product; and production of standard curves with R-squared values higher than 0.98. At the end of the course, each student was able to perform qPCR as well as RNA isolation, cDNA synthesis, primer design, and primer validation with sufficient accuracy to independently generate the expression profiles presented in this report, a skill that is transferable to many laboratory settings. Students with this advanced molecular biology skill are technically qualified for positions in molecular diagnostics measuring viral or bacterial loads and genetic mutations, forensics, the food industry, agriculture, and drug development (Valasek and Repa, 2005).
Pre- and Posttesting
Pre- and posttests were administered during two semesters to assess the students’ skills and confidence in molecular biology topics and techniques. Twenty-seven students performed a self-assessment in forced-choice questions and then in complementary open-ended questions (Supplemental Figures S1 and S2). Pre- and posttests were unannounced, administered at the beginning and end of each semester, and evaluated for each student individually.
The Python Project Online Survey
A 19-question online survey was administered via SurveyMonkey to 82 former students of the Python Project who had taken the course between 2009 and 2013 (exempt status, protocol 13-0217; Figure S3); 41 former students responded over the 2-wk data-collection period, email addresses for six students were not valid, and 35 did not respond. Questions required either Likert scale or open-ended responses. Participants were given 2 wk to complete the survey and were informed that completion of the survey was voluntary and without compensation. They were also informed that all responses were anonymous and would in no way affect current or future relationships with the University of Colorado at Boulder.
Statistical Analyses
An unpaired Student's t test was performed on pre- and posttest results with a p value of < 0.05 considered statistically significant.
RESULTS
Student Research Projects Generate Novel, Meaningful Data
Validation of Student Experimental Design and Technique.
Because of the challenges associated with working with the python genome, which has not yet been annotated, students are required to repeatedly validate their results throughout the semester (Table 5). The whole-genome shotgun (WGS) sequence of Python molurus bivittatus was used in the semesters reported here (National Center for Biotechnology Information [NCBI] accession AEQU0000000.2, genome v1.0 and v2.0; Castoe et al., 2013). Students search the python WGS using the transcript sequence of their gene of interest obtained from a closely related reference species, typically chicken (Gallus gallus) or anole lizard (Anolis carolinensis). Contiguous DNA sequences (contigs) from the python WGS that align with the gene in the reference species with greater than 80% identity are assembled to create the putative Burmese python transcript. As contigs that align with the gene in the reference species are identified, students highlight regions of the reference species transcript and use the order of highlighting to assemble the contigs into the putative Burmese python transcript (Figure S4A). In silico validation of this assembled transcript is achieved by translating the sequence (http://web.expasy.org/translate) and by searching protein databases using the Protein Basic Local Alignment Search Tool (Standard Protein BLAST, http://blast.ncbi.nlm.nih.gov; Figure S4B). If the target protein is returned, primers are designed using Primer3, an open-source software package (http://primer3.sourceforge.net). A second in silico validation is performed by using Primer BLAST to determine whether the primer specifically amplifies the gene of interest (Figure S4C).
| Technique | Validation | Criteria for success |
|---|---|---|
| RNA isolation | 1. Spectrophotometry | A260/280 greater than 1.8 |
| 2. Denaturing gel electrophoresis | Two clear bands representing 18S and 28S rRNA | |
| Primer design | 1. Amino acid translation of assembled transcript | One reading frame with no stop codons introduced |
| 2. In silico Protein BLAST | Correct protein returned in a closely related species | |
| 3. Conventional PCR | Correctly sized product amplified | |
| 4. qPCR | Single melt curve for the amplified product | |
| cDNA synthesis | Conventional PCR | Amplification of a housekeeping gene in all samples |
| Measurement of gene expression | Multiple qPCR experiments | Results must be repeatable |
In vitro validation of primers is achieved by amplifying cDNA synthesized from Burmese python liver or heart RNA using conventional PCR. The desired PCR product can be determined by size on a 1.5% agarose gel (Figure S5). Finally, the primers are examined using qPCR to determine whether the product is amplified at an acceptable cycle threshold, indicating the abundance of the gene, and whether a single product is amplified with the expected melt temperature (Figure S6). In semesters when time allowed, the PCR product was cloned (TOPO PCR Cloning, Life Technologies, Grand Island, NY) and sequenced (ACGT, Wheeling, IL) to validate the sequence of the expected PCR product and revealed that PCR using the designed primers amplified the predicted product sequence.
Validation of Student Data.
A final important validation is the reproducibility of results both from semester-to-semester and in other species. For example, in Fall 2010, students generated novel gene expression data for 16 genes hypothesized to be involved in proliferative processes in the python liver during digestion. Because the course was designed to create continuity between semesters and to coordinate with progress made in the sponsor's laboratory, the project evolved from semester to semester, and validation of one semester's data was often performed by students in the next semester. Studies performed in the sponsor's lab revealed that growth of python tissue after feeding may be attributed to components of the serum (Riquelme et al., 2011). In Fall 2011, we therefore extended the Python Project studies to examine the effects of serum obtained from pythons at different postprandial days on gene expression in cultured mammalian (rat) liver (hepatoma) cells; fatty acids present in serum isolated from postprandial python have been shown to induce extreme physiological growth of mammalian cells (Riquelme et al., 2011). A strategy similar to the one outlined in the previous section was utilized, except cloning was not performed, because transcript sequences were available for all genes studied in the rat. The gene expression studies were repeated using the same set of genes to examine both the direct effects of serum on hepatocytes independent of other cell types and the tissue milieu and to extend the examination of these factors to mammalian cells. Although three genes were not expressed in cultured liver cells, expression levels for the remaining 13 genes correlated with expression in the python liver, supporting the reproducibility of data across species despite the challenge of designing primers in an unannotated genome and measuring expression in the python.
In the semesters that followed, the students’ experiments paralleled newer ideas generated in the sponsor's laboratory. Expression of genes related to metabolism was examined to complement expression of growth-related genes obtained from hypertrophic heart and liver. In Fall 2013, students examined genes involved in cardiac regression that occurs in the Burmese python heart at 6 d after feeding. Regression of cardiac hypertrophy is a largely understudied area that may provide the sponsor's laboratory with valuable information about the mechanisms that contribute to tissue atrophy in human disease. And in Spring and Fall 2014, students measured expression of genes related to mitochondrial biogenesis and adipogenesis, respectively.
Students Acquired Laboratory Experience and Increased Confidence in Their Ability to Perform Independent Scientific Research
Grading of Students.
Throughout the course, students developed research projects that produced novel data and required the presentation and defense of their choices of gene and hypothesis. Students also prepared summaries of the results in scientific paper format and presented their data in a public poster presentation. As such, assessment was in part based on knowledge of the rationale and methods used and on scientific reading, writing, presentation, and documentation of experimental methods and results. Students were encouraged to interact with instructors and other students in a lab meeting format to convey information related to their experiments and to ask for advice on problems with their experiments. Grading was therefore weighted as follows: midterm written exam (20%; Figure S7); literature review (10%); final research report (Figure S8), poster presentation, and laboratory notebook (20%); and quizzes, worksheets, and participation, including a 10-min oral presentation (20%). The final research report was submitted at the end of the semester in place of a final examination. Students received edits from the instructors on their literature reviews and were instructed to include them in the introduction section of the final research report. Students also benefited from receiving feedback on their research during the poster presentation from members of the MCDB Department before they submitted their final research reports.
Assessment of Students’ Gain in Technical Abilities and Data Production.
The Summer Undergraduate Research Experiences (SURE) survey was used as a model to develop the pre- and posttests administered to students (Lopatto, 2004). Modifications were made by necessity because of the nature of the course. For example, peer mentoring and a summer program component were not included in the Python Project. However, open-ended questions focused on similar topics, including experience in labs and role in the project and postgraduation plans both before and after involvement with the course. Forced-choice questions focused on self-evaluation of research lab skills.
Students were asked to assess their confidence in their laboratory research and their ability to perform molecular biology tasks in the laboratory in a written survey at the beginning of the Fall 2011 and Fall 2013 semesters. All questions were forced-choice to reflect the student's confidence in the technique (Figure S1). All students had been exposed to these techniques in other molecular biology courses before entering the course. At least 50% of students surveyed reported being “very confident” about presenting data in written form, working as a member of a group, reading and understanding scientific literature, and using resources to obtain information about scientific techniques. (Figures 2 and S1). In a posttest administered at the end of the semester, students uniformly reported improved confidence in all areas surveyed. The greatest increases on average were measured in the areas in which the students expressed the least amount of confidence at the beginning of the semester. For example, students reporting being “very confident” about troubleshooting problems during scientific experiments, which increased from 12 to 32%, and with interpreting data from techniques and drawing logical conclusions, which increased from 18 to 54% (Figure 2, open bars). These data suggest that participation in an independent research project from beginning to end was successful in improving the perceived skill sets of students. Notably, the largest average increases in perceived knowledge were noted in the technical self-assessment (Figure S1). Interestingly, when scores for individual students were compared, student learning gains were higher. This result is reflective of the lowest-scoring students in the pretest experiencing the largest increases in confidence (Figure 2, gray bars). We also observed a nonsignificant trend toward students overestimating their skills at the beginning of the semester when compared with results of open-ended questions described below, consistent with the Dunning-Kruger effect (Kruger and Dunning, 1999). Recently, the relationship between the Dunning-Kruger effect and biological sex has been measured in a small number of studies involving introductory biology and chemistry courses, with males having a higher perception of their abilities than females (Lauer et al., 2013; Pazicni, 2014). We are currently examining more closely how performance and perception are impacted by biological sex in the setting of an upper-division research-based course such as the Python Project.
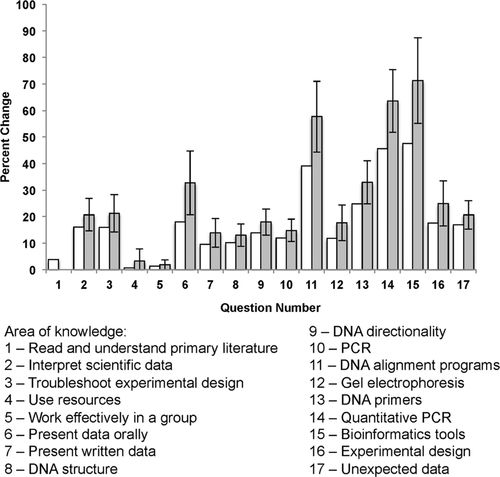
Figure 2. Pre- and posttest student self-evaluation summary. The students evaluated their confidence in seven laboratory skills (questions 1–7) and 10 molecular biology techniques (questions 8–17). (For a list of the questions, see Figure S1.) An unannounced pretest was administered on the first day of class, and an identical survey was administered on the last day. Responses were recorded for three semesters (n = 27 students). Results are reported for the average gains in each area for all students (white bars) and for individual students (gray bars). Error bars represent SEM.
Because of the unique format of the surveys administered, it was not possible to directly compare the outcomes of the Python Project with outcomes of other research courses. However, the Python Project surveys revealed similar trends to those observed in SURE survey (Lopatto, 2004), which reports students’ assessments of learning gains from participation in a variety of different research experiences. Students reported that they experienced the highest gains in their personal readiness for engaging in scientific research and in their technical abilities in the laboratory (Lopatto, 2004). Similarly, students in the Python Project reported the largest increases in confidence with skills related to performing scientific research (“interpret data from techniques and draw logical conclusions” and “troubleshoot problems during scientific experiments”), in agreement with other qualitative interview-based studies (Seymour et al., 2004). Beginning in Fall 2014, students enrolled in the Python Project will be administered the Classroom Undergraduate Research Experiences survey (www.grinnell.edu/academics/areas/psychology/assessments/cure-survey) to directly compare the outcomes of the course with outcomes of other course-based research experiences.
Students also indicated confidence in extending the knowledge to other settings outside the classroom. After students were taught to use techniques such as designing DNA primers, PCR, and gel electrophoresis, gains in the percentage of students reporting this ability were 65, 52, and 39%, respectively. Evidence for this was demonstrated during Spring 2014, when students were asked to design primers using the mouse genome, which is annotated (see Worksheet IV in the Supplemental Material); 88% of students were able to successfully design primers and validate them in silico without explicit instructions describing the procedure.
In light of positive self-reporting, we were also interested in whether the basic molecular biology skills improved from the beginning to the end of the semester. An unannounced pretest was administered on the first day of class. Open-ended and multiple-response questions addressing basic molecular biology questions on DNA structure, sequence, and conservation scored most highly, while questions on experimental techniques scored lower. When an identical unannounced assessment was administered at the end of the semester to determine whether practical experience in the skills had improved their knowledge, the students had improved in each area of basic molecular biology tested. The largest gains were measured in spectrophotometry, which is required to measure DNA concentrations, and PCR protocols; scores were increased by 42.9 and 64.3%, respectively (Table 6).
| Pretest average score | Posttest average score | Percent increase in score | |
|---|---|---|---|
| Gel electrophoresis | 78.6 | 92.8 | 14.2 |
| DNA structure | 80.7 | 85.7 | 5 |
| Reverse complement DNA sequence | 85.7 | 100 | 14.3 |
| Parameters affecting annealing temperature | 14.3 | 78.6 | 64.3 |
| Properties of Taq polymerase | 64.3 | 92.8 | 28.5 |
| Spectrophotometry | 57.1 | 100 | 42.9 |
| Define “conserved sequence” | 71.4 | 92.8 | 21.4 |
Students Expressed a Positive Attitude toward Research at the End of the Python Project and Maintained That Opinion Several Years
Student comments were collected by the university as part of a faculty course questionnaire (FCQ) and were categorized by the authors as follows: opinion of 1) the course, 2) instructors and teaching style, 3) structure of the course, and 4) the course compared with other laboratory courses in the same department. Out of 58 students, 54 (93%) surveyed between Fall 2010 and Spring 2014 completed FCQs. Of these student respondents, 57.4% (31/54) also provided comments, and 89% of the comments fell into the four categories. Fifteen percent of students provided statements about their overall opinion of the course, which was overwhelmingly positive. Twenty-two percent of student comments reflected a positive opinion of the course compared with other laboratory courses in the department and emphasized the practical nature of the course: “I have learned more applicable knowledge and skills this semester in this course than the rest of my MCDB courses combined. The Python Project helped me become confident in my laboratory techniques, presenter skills, and my ability to transition into the work force” and “I had a great experience in the course—it was very challenging, but I feel like I learned a huge amount. I also feel like the skills I’ve learned through the class are pretty unique and they will help me in the future when I am applying to graduate schools.”
These positive opinions are further supported by answers to forced-response FCQ questions at the end of each of eight academic terms, relative to the mean scores for all courses. A total of 109 FCQs were distributed, and 96 (88%) were collected by university representatives. Scores were consistently higher than the averages for other upper-division natural science courses (Figure S9). For example, the Python Project consistently scored higher on a 0–6 scale (0 indicating “lowest” and 6 indicating “highest”) compared with other courses in the university. Over eight semesters, students in the Python Project rated the course 5.71 for “course overall” compared with 4.8 for all courses offered at the university and 5.66 for “How much you learned” compared with 4.8 for university courses. However, because not all students responded to this voluntary questionnaire, we cannot make conclusive statements about whether sampling accurately represents the population of students who took the course. In the future, to increase the response rate, we will administer the survey in class while also ensuring respondent anonymity by having an unrelated member of the faculty administer the survey without the instructors present.
Importantly, the positive opinions of students were maintained after their participation in the Python Project. Of 82 former students, 41 responded to an online survey administered in Spring 2014. More than 90% of respondents who had taken the course between 1 and 4 yr earlier reported that they agreed or strongly agreed with the statement “I had an overall positive experience in the Python Project laboratory course” (Figure 3A). When asked to compare their experience with other laboratory courses they had taken, 95% of respondents reported that they agreed or strongly agreed with the statement “I prefer the structure of lab courses similar to the Python Project” (Figure 3B). Students further reported acquiring more critical-thinking skills because of the Python Project (88%), and they enjoyed the relationship between the course and the research performed in the sponsor's laboratory (97%; Figure 3, C and D).
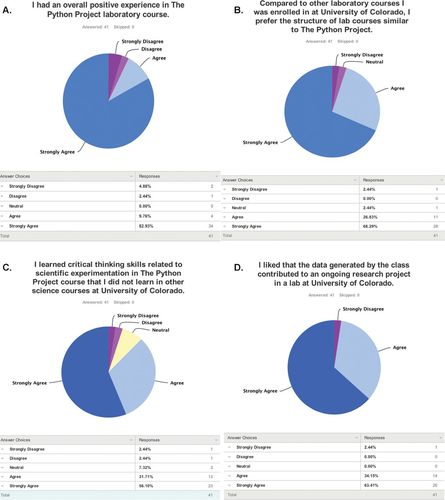
Figure 3. Student opinions of the Python Project. An annonymous survey was administered through SurveyMonkey to students who had completed the course between 2009 and 2013. The percentage of responding students in each category is indicated below the circle graphs.
Participation in the Python Project Promoted Retention in Science Careers
In addition to assessing the success of the Python Project based on the quality and reproducibility of data generated by students, we were interested in how the course shaped their careers and education plans. A survey was administered at the beginning and end of the semester during Fall 2010, Fall 2011, and Fall 2013 to ask whether students entered the course with an interest in pursuing science careers and whether participation in the course influenced their plans. Forty-two percent of students entering the course expressed an interest in pursuing a PhD in the field of biology, while 30% of students indicated an interest in applying to medical school. Approximately 20% of the students were undecided. By the end of the semester, 11% more students indicated a desire to apply to a PhD program in biology, and 22% fewer students were interested in medical school (Figure 4). When students were considered individually, the greatest shift was observed in the seven students who were undecided at the beginning of the semester; four indicated an interest in graduate school in the biological sciences, and two reported that they intended to seek a position as a research assistant. An additional three students who had planned at the beginning of the semester to seek research assistant positions had shifted their plans to graduate school in the biological sciences at the end of the semester.
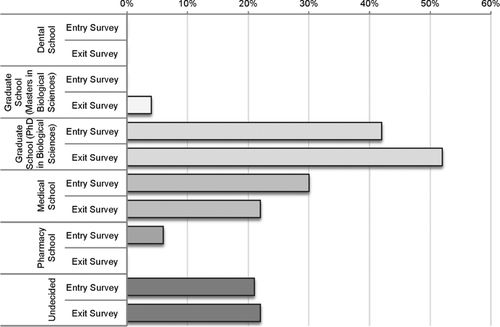
Figure 4. Intended education plans at the beginning (Entry Survey) and end (Exit Survey) of students participating in the Python Project between 2010 and 2013.
Indeed, in the last two semesters taught, 92% of students in their third year of undergraduate education sought opportunities to perform research after taking the course; for example, from the Fall 2011 class, three students participated in independent study projects in the sponsor's laboratory, two students obtained research technician positions in other faculty laboratories, and one student enrolled in two semesters for credit to extend his studies on the python. Similar trends were observed after each semester. In Fall 2011, 87.5% of students interested in laboratory research secured volunteer or independent study positions in the MCDB Department while enrolled in the Python Project. In Fall 2013, 80% (10/16) of students interested in securing either volunteer research positions or independent study credit were successful. These data are consistent with other studies that demonstrate research experience fosters retentio–n in the biosciences (Hunter et al., 2007; Russell et al., 2007).
To extend these short-term analyses, we administered an online survey to students enrolled between 2009 and 2013 to determine whether the Python Project contributed to retention in science, technology, engineering, and mathematics education and careers. Of 82 students, 41 responded to the survey. Because survey answers were anonymous, no information about the respondents was provided, except the semester in which they participated in the course. Seventy-five percent of students who responded to the survey agreed or strongly agreed that their decision “to engage in independent research was positively influenced by my participation in the Python Project” (Figure 5A), and 95% of respondents agreed or strongly agreed with the statement “My participation in the Python Project increased my understanding of performing novel research” (Figure 5B). In agreement with these statements and the percentage of students who had secured research positions while taking the Python Project, ∼68% of former students reported working in a research laboratory during their undergraduate education and after their participation in the Python Project (Figure 5C).
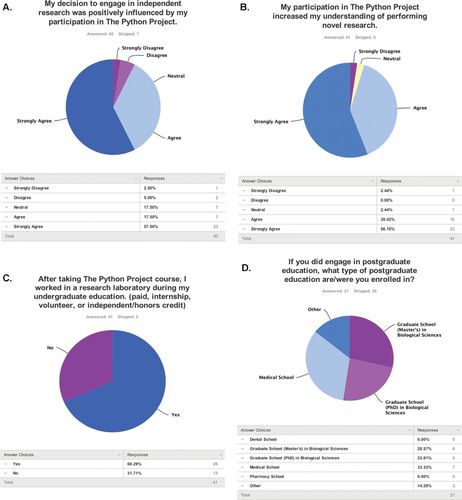
Figure 5. Effect of the Python Project on students’ career and education choices.
Seventy-five percent of respondents had completed their undergraduate education. Interestingly, of respondents who had graduated, 71% had pursued postgraduate education: 26% pursued a master's degree in biological sciences, 24% worked toward a PhD in biological sciences, 33% attended medical school, and 14% indicated “other” postgraduate education (Figure 5D). Of the remaining 32.3% graduates who did not pursue postgraduate education, 46.7% were employed as research assistants in an academic or pharmaceutical research laboratory. An important caveat to interpretation of these data and data obtained from similar voluntary surveys is that the information may be skewed due to the population of responding students.
Recommendations for Implementation
From our experience with the Python Project, we make the following general recommendations for implementing similar research-based undergraduate courses:
Unique model organisms: It is important to emphasize that although we believe that part of the success of the course is attributable to the unique animal model used, there are many genomes that have been sequenced but not annotated that would benefit from the same types of studies. Genome sequencing has becoming relatively inexpensive; however, annotation algorithms continue to return up to 50% of the gene as “conserved hypothetical genes.” There are currently more than 1000 bacterial and 4000 eukaryotic sequencing projects in progress, most of which are WGSs that will be made publicly available (www.genomesonline.org). These leave open the possibility of many inexpensive models in which to replicate the gene annotation performed as part of the Python Project.
Small class size: Although the course-based research experience does not provide 1:1 interactions with a researcher as apprenticeship-style approaches do, it does provide a higher faculty-to-student ratio than is normally offered at a large university. The benefit of such interactions in our experience and in our analyses is a more rewarding personal experience for both the instructor(s) and students. A challenge to creating an environment that promotes personal relationships and encourages students to express opinions and ideas is the high level of investment on the part of instructors to maintain trust and respect.
Modification of techniques utilized: This model could be cost-prohibitive in its current format, which uses qPCR; course expenses (including all laboratory equipment) distributed over the total number of students (120) in eight semesters was approximately $280 per student. However, creative adaptation of this model is feasible if a department commits financial support for several years. For example, departmental funding was not provided for instructors’ salaries; commitment of a university department would help offset the overall cost of the course. Additionally, shared departmental resources may abrogate the need to purchase large equipment for the class. Finally, semiquantitative PCR, which requires a conventional thermal cycler, could be used in place of a qPCR thermal cycle and would drastically reduce the cost of reagents and disposables.
Involvement of the sponsor's laboratory: An important consideration in the implementation of this model is the relationship between the course and the sponsor's laboratory; the students benefit from the research experience while the course brings undergraduate students to the laboratory, so more senior scientists who are not involved in teaching can interact with students. Additionally, the mentor laboratory benefits from the data generated by the students. As with the Python Project, students can engage in measuring expression of a large number of genes, or this model could be extended to genetic screens or identification of drug targets or candidates. Alternatively, a more global approach can be taken through participation in crowdsourcing projects such as the Small World Initiative with oversight from a mentor laboratory interested in similar subjects.
The sense of membership in a research department could be conferred through guest lecturers presenting their own research that is related to a curriculum topic, for example. The idea of membership in a bigger research effort could also be experienced through continued student involvement in the course. For example, in the last two semesters of the Python Project, former students have served as guest lecturers to present a basic principle or technique or to demonstrate an oral research presentation or poster. This provides current students with a sense of community as well as providing former students a feeling of continuity with the project. We have also observed that students often are under the impression that they are members of the mentor lab; identifying with the mentor laboratory could be an indicator of retention in science; however, further analysis is required to specifically test this hypothesis in this setting.
Utilization of student data: Although the amount of data contributed by the students may be significant to a laboratory interested in preliminary data for a larger project, an authentic research experience should ideally include involvement in development of a peer-reviewed publication. To address requests from students for continued involvement in their research and in response to recommendations that undergraduate research experiences have broader relevance (Auchincloss et al., 2014), in Spring 2014, the instructors piloted a project in which students prepared a single manuscript that summarized the research of the entire group with the goal of receiving feedback from students on how to best implement this extended project in future semesters. Students were enthusiastic about the project; all 16 enrolled students volunteered to participate over the summer with the understanding that they would not receive credit or be paid for their effort. Based on the quality of the final papers at the end of the semester, the instructor selected two students to lead the project. These two students met with the instructor weekly throughout the summer to develop the manuscript. All 16 students who were enrolled in Spring 2014 and who contributed gene expression data were kept apprised of the progress, and volunteers were selected throughout the process to edit sections of the text. As such, students are exposed to responsible conduct in publishing research and the peer review process. Currently, we are in the final stages of preparation and have plans to submit the manuscript to the Journal of Student Research, a peer-reviewed journal that publishes research by undergraduate and graduate students. All 16 students are included as authors on the manuscript. Based on the feedback from this pilot project, we aim to offer a second semester of the Python Project that allows students who are interested in preparing a publication based on their work in the previous semester. Although we are uncertain of the exact outcomes of this extended project at this time, we believe it will further enhance the experience for the students.
Inclusion of this type of experience is made possible by many quality peer-reviewed undergraduate journals. We are aware of compelling criticisms of undergraduate journals that put forth valid arguments, including the fact that publication in a student journal precludes publication of the data in a mainstream scientific journal. We focused on a student journal for this pilot project based on retrospective assessment of the cohesiveness of the data obtained from the semester. In the future, we will plan each semester to test a specific overriding hypothesis with the intention of submitting a publication to a mainstream scientific journal. Students will form hypotheses about their specific gene(s) and also about how their genes fit into the larger hypothesis of the class.
CONCLUSIONS
In light of the outcomes measured, this model successfully provided 133 students with an authentic research experience and met the goals outlined at its inception. Students obtained reproducible data both in their own experimental replicates and in comparison with data obtained from previous semesters and in the sponsor's laboratory. These data were then used in the sponsor's laboratory to guide the research project on organ growth in the Burmese python. For example, preliminary data in the sponsor's lab suggested that many genes related to lipid metabolism are important to organ growth in the python. Student data obtained in Fall 2011 revealed that, indeed, many of these candidate genes are differentially regulated with the feeding cycle of the python. Finally, considering that the majority of students continued their science education or chose to work in a laboratory, the Python Project was also successful in encouraging students to remain in the field of science. We therefore recommend this model to other universities seeking an efficient means for extending research opportunities to a larger number of students than is afforded by independent study and summer programs.
ACKNOWLEDGMENTS
We thank Jennifer Knight for critical review of the manuscript and Bailey Keller for assistance with survey design. Development of this article was supported by an HHMI Professorship awarded to L.A.L. Funding for the Python Project course was provided by the HHMI and Amgen.



