An Imaging Roadmap for Biology Education: From Nanoparticles to Whole Organisms
Abstract
Imaging techniques provide ways of knowing structure and function in biology at different scales. The multidisciplinary nature and rapid advancement of imaging sciences requires imaging education to begin early in the biology curriculum. Guided by the National Institutes of Health (NIH) Roadmap initiatives, we incorporated a nanoimaging, molecular imaging, and medical imaging teaching unit into three 1-h class periods of an introductory course on ways of knowing biology. Activities were derived from NIH Roadmap initiatives in nanomedicine, regenerative medicine, and nuclear medicine. The course materials we describe contributed positively to student learning gains in quantifying and interpreting images, in characterizing imaging methods that provide ways of knowing biological structure and function, and in understanding scale in biology and imaging. The NIH Roadmap provides a useful context to educate students about the multidisciplinary imaging continuum.
INTRODUCTION
Currently, undergraduate science courses are not keeping pace with research advances and are lacking an intellectual continuum that fosters quantitative, multidisciplinary studies (Bialek and Botstein, 2004). Curriculum reform at the predoctoral level is especially needed in biology and its integration with the imaging sciences (Sullivan, 2000) because of the rapidity of imaging advances and multidisciplinary expansion of imaging applications (Paschal, 2003). With educational opportunities being created by neuroinformatic endeavors like the Human Brain Project (Shepherd et al., 1998), there is a need to integrate imaging into the undergraduate biology curriculum to develop information technology skills for all students and provide the necessary foundation for those biology students interested in taking advanced imaging courses (Hurd and Vincent, 2006). The demand for multidisciplinary imaging skills is high in the marketplace (Smaglik, 2005), and the imaging infrastructure has been consolidated by formation of the National Institute of Biomedical Imaging and Bioengineering (Hendee et al., 2002) and the National Institutes of Health (NIH) Roadmap initiative (Fee and Pettigrew, 2003; Zerhouni, 2003). The NIH Roadmap (http://nihroadmap.nih.gov/) was designed to accelerate research and medicine based on three major themes: New Pathways to Discovery, focused on improving our understanding of biological systems; Research Teams of the Future, which explores alternative models to organize research teams; and Reengineering the Clinical Research Enterprise, which aims to better integrate current research infrastructure and improve clinical outcomes assessment. This course was structured around the initiative within the New Pathways to Discovery theme: to advance nanomedicine, tissue engineering and regenerative medicine, and nuclear medicine using a variety of imaging techniques.
The overarching learning goals of an imaging teaching unit early in the introductory biology curriculum are that students understand the integrated landscape of imaging science requires preparation through multidisciplinary studies and that students are better prepared for this rapidly advancing field whether as consumers or future providers of molecular medicine (Dzik-Jurasz, 2003).
Biological imaging is a technique that can bring quantification into biology education and offers new insights into biological structure and function through visualization of processes at the nano, molecular, cellular, and system-level scale. Innovations in biological imaging require translation from the laboratory bench to the undergraduate, graduate, and medical classroom. However, it is challenging to provide a context in the undergraduate curriculum that will highlight the multidisciplinary scope of imaging science and its integrated continuum with biology. Furthermore, instruction in biological imaging must be able to overcome student misconceptions (Table 1), to engage students with content that is interesting to all students but requires little background knowledge, and to encourage learning gains that are self-directed within an active-learning framework. Here, we introduce a teaching unit on biological imaging developed for three 1-h class periods in an introductory undergraduate biology course, Ways of Knowing Biology (WOK). We show that our course design, which drew from NIH Roadmap initiatives, was significantly associated with student learning gains in understanding key concepts and developing core skills in imaging.
| MisconceptionsStudents mistakenly think that: | |||
| -Biological science and imaging science are unrelated | -Biological images can only reveal structure but not function | -Biological images are pretty pictures but not quantifiable | -The impact of imaging techniques on biology and medicine is limited |
| Learning GoalsStudents will: | |||
| -Conclude that the scale of biology studied must match the scale captured by an imaging device | -Appraise the utility of imaging tools to examine structure and function of biological processes | -Discover that imaging is a quantitative tool used to measure data across a wide range of biological scale | -Generalize that biological images hold prominent places in advancing biology and medicine and contribute to the NIH Roadmap initiative |
| -Develop interpretation skills by examining different imaging formats and multiple biological scales using real data | -Evaluate imaging methods that provide ways of knowing structure and function | -Develop computer and computation skills | -Understand that imaging is an integrated multidisciplinary field |
| Learning OutcomeHaving achieved the learning goal, students will be able to: | |||
| -Summarize the relationship between biological and imaging scale | -To predict the rabbit Alba's color from fluorescence emission of eGFP | -Proficiently use NIH ImageJ software to calculate NanoBucky's height and image intensity | -Value that (1) nanoimaging advances the development of nanodevices and nanomedicine |
| -Accurately interpret:(1) nanoimages of NanoBucky (2) molecular images of fluorescent probe intensity from (a) bovine pulmonary artery endothelial cells (b) eGFP labeled human embryonic stem (ES) cells (3) system-level images of (a) eGFP rabbit Alba (b) MRI image of brain (c) CT image of Phineas Gage's skull | -Evaluate the intensity of eGFP labeled ES cells to understand ES function | -Apply ImageJ principles to: (1) Interpret the intensity of eGFP labeled cells | (2) molecular imaging advances tissue engineering and regenerative medicine |
| -Assess functional brain activity using 18-FDG PET measures of glucose metabolism | (2) Quantify and localize brain function using PET images | (3) system-level imaging (e.g., PET) advances nuclear medicine | |
| -To explain and give examples of imaging tools | -Identify disease using X-ray images of lung with and without pneumonia and PET images of brain with and without a tumor | ||
| -Compare 2D, 3D VR, and stereolithograph models of brain and Phineas Gage's skull and contrast them for visual information quality and biological utility | |||
MATERIALS AND METHODS
Scientific Teaching
We used scientific teaching and backward design approaches to create this teaching unit (Handelsman et al., 2004; Handelsman et al., 2007) for the WOK course. The main goal of WOK was to introduce 75 first-year college students to the breadth and methods of biology in a pass/fail environment. Supplemental Material for this imaging unit (including the syllabus, slides, pre/post questionnaires, Gillespie survey, SALG instrument, and imaging Internet resources) are available through the Wisconsin Program for Scientific Teaching (WPST) Digital Library website (http://scientificteaching.wisc.edu/materials/molecularbiology.htm). Specifically, we established learning goals in the context of the course and literature, we designed assessments that described student performances to indicate achievement of those goals, we developed activities to help students achieve the goals, and we evaluated the success of the materials by quantifying student learning gains.
Because imaging misconceptions develop where education does not keep up with technological advances (Hawkins and Dunn, 1996), we identified concepts in the imaging and biology literature that would help students integrate biology and modern imaging methods, that were reported as misconceptions, or that were recommended for incorporation into the biology curriculum (Sullivan, 2000; Hawkins and Dunn, 1996; Provenzale and Mukundan, 2005; Illes et al., 2006; Schnell et al., 2007). These concepts provided a basis for primary learning goals and specific learning outcomes (Table 1). We incorporated learning goals into student activities to delineate measurable criteria for assessment purposes. The activities to achieve these goals took the form of mini-lectures that progressed in scale from the nanoscale to system level scale, NIH ImageJ analysis, in-class NIH Roadmap activities, and the Gillespie (Muller et al., 2003), Student Assessment of Learning Gains (SALG; Wisconsin Center for Educational Research, 1997), and pre/postassessments.
In-class activities and course content were developed and unified around the NIH Roadmap initiatives. More specifically, assessment activities in nanoimaging, molecular imaging, and medical imaging were drawn from nanomedicine, tissue engineering in regenerative medicine, and nuclear medicine. The course activities progressed in biological and imaging scale from nano through macroscale. As a nanoimaging activity, students quantified electron microscopy images of carbon nanofibers arranged into the shape of Bucky Badger (NanoBucky) (Hamers, 2007). The molecular imaging activities involved interpreting fluorescent microscopy images of fluorescently labeled bovine pulmonary artery endothelial cells (available in NIH ImageJ), enhanced green fluorescent protein (eGFP) expressing stem cells (Zwaka and Thomson, 2005), and macroscopic fluorescent images of the eGFP rabbit Alba (Stewart, 2006). System level activities required clinical evaluation of a NIH positron emission tomography (PET) exercise (NIH, 2007) supplemented with a patient case study based on 18-FDG PET images collected at the University of Wisconsin-Madison Cyclotron and PET Research Center. In addition, students evaluated computed tomography (CT) images of Phineas Gage's skull (Ratiu and Talos, 2004) and MRI images of human brain in two-dimensional (2D) images, three-dimensional (3D) virtual reality (VR), and physical 3D formats. Virtual reality models of human brain and Phineas Gage's skull were generated and then printed in 3D as physical stereolithograph models at the UW-Madison New Media Center (Kelley, 2007). We viewed the VR brain and skull models in the virtual reality markup language (VRML) format with VRMLView (Systems in Motion; Norway).
To provide a better understanding of the actual lessons and student activities, we will provide an in-depth description and active-learning approach for the NanoBucky exercise. Students were asked to bring in their laptops for this in-class activity. Students organized themselves into groups and used the NIH ImageJ applet (http://rsb.info.nih.gov/ij/applets.html), which did not require prior installation but did require an Internet-enabled classroom. Students were provided with NIH ImageJ instructions to use NIH ImageJ applet software from the ImageJ Documentation Wiki (http://imagejdocu.tudor.lu/imagej-documentation-wiki). Students actively learned how to initiate the software, load an image, and analyze the height of NanoBucky. This activity familiarized students with nanomaterials that are being developed as part of the NIH Roadmap initiative and enabled students to understand the nanoimaging scale by calculating the height of NanoBucky based on the scale legend provided in the image (http://hamers.chem.wisc.edu/research/nanofibers/index2.htm); to understand how imaging provided scientists with ways of viewing and assessing nanomaterials and, in the future, nanodevices for quality control; and to recognize that imaging is a quantitative tool in biology by observing, measuring, and interpreting biological images. The NanoBucky activity was completed in ≈30 min.
All the course activities aligned directly with the learning goals (Table 1) and exemplified how biological images advance the NIH Roadmap initiatives. The relationship between biological scale and imaging scale was exemplified by nanoimages of NanoBucky, molecular images of fluorescent stained endothelial cells available in Image J (Abramoff et al., 2004), eGFP labeled human embryonic stem cells, the eGFP rabbit Alba, MRI images of brain, and CT images of Phineas Gage's skull. Students could understand that imaging provides ways of knowing biological structure and function by interpreting the intensity of eGFP labeled gene expression during human embryonic stem cell differentiation and images of brain function using 18-FDG PET and functional MRI. The use of MRI and CT images to create stereolithograph models of brain and skull, respectively, exemplified the broader application of images and novel visualization approaches. In addition, students could develop their evaluation skills by comparing 2D images, 3D VR, and stereolithograph models in terms of visual information quality and biological utility. Computer skills could be developed by using the NIH ImageJ software to quantify NanoBucky's height and intensity and to apply this knowledge when interpreting fluorescently labeled endothelial and stem cells. With NanoBucky, students could gain an understanding that nanoimaging of nanomaterials influences the development of nanodevices, which have applications in nanomedicine; that molecular imaging of human embryonic stem cells can inform tissue engineering, which can advance regenerative medicine; and that PET imaging with radioactive tracers can influence nuclear medicine.
Diversity
This course addressed diversity in learning styles, but it was much more challenging to simultaneously address gender, cultural, and socioeconomic diversity. Diversity in students' educational backgrounds was accounted for in this teaching unit by incorporating multiple modes of teaching and assessment activities. Mini-lectures provided background information that minimized discrepancies in educational background. Audiovisual aids were used to clarify concepts in an interesting and understandable manner. The video “Powers of Ten” introduced the concept of scale, and a movie clip of the “Hulk” illustrated fluorescence. Using NIH ImageJ, students quantified images and observed how images can be utilized using hand-held models of a brain and of Phineas Gage's skull. We accounted for different test-taking styles by using a variety of formats to assess learning gains: oral discussion, written answers, online and in-class surveys, and online pre- and postcourse assessments. To address gender and cultural diversity, the significant contributions of minority and female scientists from various countries were specifically incorporated into the mini-lectures. For example, the work of the female Polish chemist and physicist Marie Curie and English biophysicist Rosalind Franklin were highlighted for the contributions to nuclear medicine and genetics, respectively. To address socioeconomic diversity, we built upon the common campus culture held by all UW-Madison students and incorporated the work of scientists who made contributions to imaging and who were either from the state of Wisconsin or were affiliated with UW-Madison. For example, we highlighted the work of Raymond Damadian, who attended UW-Madison on a Ford Foundation Scholarship as an undergraduate and went on to invent and develop the first MRI scanner, Indomitable.
Active Learning
Students were engaged in learning actively in multiple ways. For example, visualization of biological processes using images engaged students in scientific thinking. By viewing and analyzing nanoimages, molecular images, and medical images, students could gain an understanding of the relationship between biological and imaging scale and realize that biological science and imaging science are related by scale. By viewing and analyzing images with fluorescent probes and radionuclear markers, students were encouraged to understand how images provided ways of knowing biological structure and function. When introducing different imaging topics, we selected interesting images that students could observe, quantify, and interpret. As examples, students used a freely available software program, NIH ImageJ (Abramoff et al., 2004), to quantify images and evaluated image visualization and utility by comparing 2D slices of brain and of Phineas Gage's skull with 3D VR and printed 3D stereolithograph models. Through introduction of NIH ImageJ analysis software, students could extend their conceptual understanding of imaging analysis into a technical skill using real data. By evaluating images, students understood how biological images advanced nanomedicine, regenerative medicine, and nuclear medicine and contributed to the NIH Roadmap initiatives.
Several varieties of images were used in this course module and are described in Table 1. These can be categorized into 2D and 3D images. The 2D images included (1) nanoimages of NanoBucky; (2) molecular images of fluorescent probe intensity from (a) bovine pulmonary artery endothelial cells and (b) eGFP labeled human embryonic stem (ES) cells; (3) system-level images of (a) the eGFP rabbit Alba, (b) MRI images of human brain, and (c) CT image of Phineas Gage's skull. The 2D images of human brain and Phineas Gage's skull were rendered into a computerized, 3D volume in the VRML format and displayed to students through a projector. In addition, the computerized VRML models were printed in three dimensions as physical stereolithograph models which students held in their hands.
Assessment
Through these activities, we were able to evaluate student learning gains summatively by comparing the results of a pre- and post- survey and formatively throughout the teaching unit by using assessments for active-learning activities, the Gillespie scale (Muller et al., 2003) for rating visual information quality, and the SALG assessment (Wisconsin Center for Education Research, 1997), which is an instrument developed by Elaine Seymour at the University of Wisconsin Center for Educational Research to assess student perception of learning gains as a function of course design and delivery (Seymour et al., 2000).
Summative pre- and postassessments showed that the learning gains in the unit were achieved. Students were asked to define biological scale, list and explain three imaging modalities, and quantitatively interpret X-ray images of lung with and without pneumonia. To assess learning gains, a rubric was established to code responses with a 3-point scoring scale with 1 = incorrect/unknown, 2 = a basic understanding, 3 = an advanced explanation using quantitative terminology.
In-class assessments included group discussions, activities, and evaluations that indicated to teachers, as well as students, how well concepts were understood. Here we report technical gains in the ability to use the NIH ImageJ program to quantify biological images in partial fulfillment of our learning goals. Responses from student groups were collected and compared with repeated measures by the instructor. Student proficiency with ImageJ software was assessed with a t-score. We used the Gillespie rating scale (Muller et al., 2003) to assess students' perception of biological image models relative to a baseline model to examine whether students made an association between image visualization and utility in biology. In our assessment, 2D images were used as the baseline for assessing VRML and stereolithograph models for the quality of their visual information and for their biological utility. Gillespie ratings were coded on a 4-point Likert scale (1 = Inferior, 2 = Similar/Equivalent, 3 = Superior [similar information more rapidly assimilated], 4 = Superior [additional information provided]). Students also completed our implementation of the SALG instrument, which has been successfully administered to assess student learning (Anderson, 2006; Casem, 2006). Responses to the SALG were anonymous and collected online using Zoomerang software (MarketTools). Categorical responses were coded on a 5-point Likert scale (0 = Not applicable; 1 = Not at all, 2 = A Little, 3 = Somewhat, 4 = A Lot, 5 = A Great Deal). To clarify the choice of Likert scale coarseness, a 3-point scale was arbitrarily used for pre-post assessment, whereas the Gillespie and SALG ratings used the historically recommended 4- and 5-point scale, respectively. Questions in the student gains category focused on the gains in learning goals for this imaging course, and the course design questions were centered on engagement activities and course content. The association between student gains and course design was used to assess alignment between student learning gains and the activities that were developed to help students achieve the learning goals (Kelley and Johnson, 2007). The Gillespie associations and SALG associations were determined using polychoric (Fox, 2004) correlation software in R (Development Core Team, 2005). The polychoric correlation, rho, is a useful statistic to understand associations in categorical data and is preferred to the Spearman correlation because the discretizing latent variable thresholds are estimated (Wallenhammar et al., 2004). Two-tailed significance was assessed after a rho to t conversion on N-2 degrees of freedom. A corrected p value < = 0.05 was considered significant. Analyses were conducted in R version 2.4.1 and SPSS version 14.0.
RESULTS
Summative Assessments
The distribution of the pre- and postassessment learning scores for biological scale, imaging tools, and image interpretation are displayed in Figure 1. The Wilcoxon signed rank test on pre- and postassessment learning scores indicate that students made significant learning gains in interpreting images (Z = −2.81; p = 0.005; n = 34), distinguishing imaging modalities (Z = −3.70; p = 0.00; n = 36), and understanding biological scale (Z = −4.64; p = 0.00 [2-tailed]; n = 35).
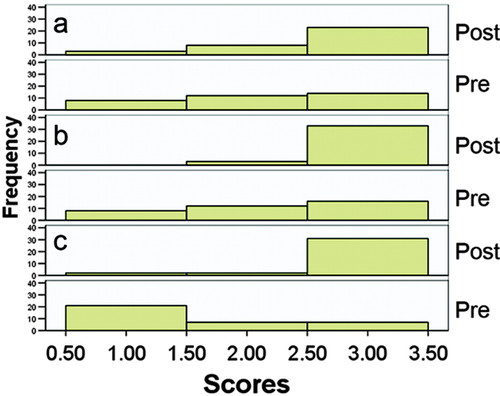
Figure 1. Summative assessment of image interpretation, imaging tools, and biological scale. Histogram of responses to pre- and post- course assessments focusing on learning goals in which students were asked to (a) interpret images of healthy and unhealthy lungs (n = 34), (b) explain and give examples of imaging tools (n = 36), and (c) define biological scale (n = 35). The change in distributions between the pre- and postcourse assessment indicated that students made positive learning gains toward our learning goals, which were for students to understand the prominent place of quantitative imaging in medicine, to recognize the scope of imaging tools and their application to examine structure and function, and to understand the meaning of biological scale in imaging.
In-Class Assessments
Students quantified the height of NanoBucky using NIH ImageJ (Figure 2). Groups of students reported NanoBucky's height in micrometers (mean ± SEM: 31.9 μm ± 0.49, n = 3) and pixels (mean ± SEM: 342.9 pixels ± 2.0). When compared with instructor estimates for NanoBucky's height in micrometers (mean ± SEM: 32.2 μm ± 0.25, n = 10) and pixels (mean ± SEM: 337.7 pixels ± 2.7, n = 10), the height estimates in micrometers [t(11) = −0.52; p = 0.61; unpaired, 2-tailed] or pixels [t(18) = 1.56; p = 0.14; unpaired, 2-tailed] were not significantly different between the students and instructor.
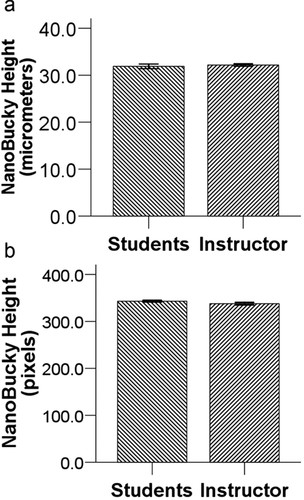
Figure 2. In-class assessment of image quantification. Measurements of NanoBucky's height using ImageJ were reported as (a) micrometers (n = 3 for students; n = 10 for instructor) or (b) pixels (n = 10 for students; n = 10 for instructor). Students demonstrated proficiency in using imaging software because the height estimates in micrometers [t(11) = −0.52; p = 0.61; unpaired, 2-tailed] or pixels [t(18) = 1.56; p = 0.14; unpaired, 2-tailed] were not significantly different between the students and instructor. This satisfied our learning goal for students to recognize imaging as a quantitative tool.
The Gillespie ratings indicate that students perceived a strong association between the quality of visual information contained in images and their biological utility (Figure 3A). After multiple comparison correction (Figure 3B), significant associations were positive except for a negative association between the visual information contained in the stereolithograph of Phineas' skull and the general utility of VRML models in biology (Table 2). This indicates students made evaluations across the different model types and identified a tradeoff in utility between VRML and stereolithograph models for biological visualization. This may be attributable to the instructor demonstrating that the virtual reality model permitted navigation of structures internal to the skull as students took the perspective of the tamping iron that passed through the Phineas Gage skull when the instructor used the zooming tool to navigate within the cranial vault from superior to inferior and vice versa. Students could not physically navigate the stereolithograph model in the same way, which may have produced the negative polychoric correlation.
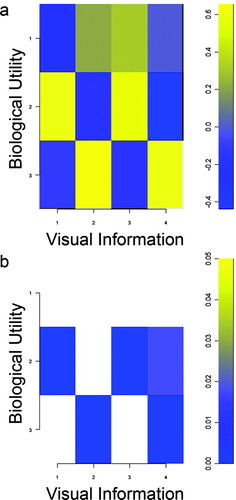
Figure 3. Gillespie associations of visual information with biological utility. (a) Polychoric correlations of biological utility (1 = 2D, 2 = VRML, and 3 = stereolithograph) with visual information (1 = brain VRML, 2 = brain stereolithograph, 3 = Phineas VRML, 4 = Phineas stereolithograph) were both positive and negative and indicated that students evaluated VRML and stereolithograph models. (b) P values of significance (p ≤ 0. 05, corrected for multiple comparisons) for polychoric correlations in Figure 3A. Students made positive associations between the biological utility of these models with the quality of their visual information. Only the Phineas stereolithograph had a significant negative association with VRML utility, which indicated that students made comparisons across VRML and stereolithograph model types and recognized a tradeoff between visual information and biological utility.
| Question (VI, BU) | Visual information | Biological utility | Polychoric correlation | df | P value (corrected) |
|---|---|---|---|---|---|
| 1,2 | Brain VRML | VRML | 0.61 | 48 | 0.00 |
| 3,2 | Phineas VRML | VRML | 0.57 | 48 | 0.00 |
| 4,2 | Phineas stereolithograph | VRML | −0.44 | 48 | 0.02 |
| 2,3 | Brain stereolithograph | Stereolithograph | 0.65 | 48 | 0.00 |
| 4,3 | Phineas stereolithograph | Stereolithograph | 0.60 | 48 | 0.00 |
The SALG instrument identified aspects of course design that contributed to student learning gains (Table 3). Significant associations between learning gains and course design (Figure 4A) after correcting for multiple comparisons (Figure 4B) were positive and involved the primary learning goals that were key features of our backward design (Table 1).
| Question | Topic | Mean | SD | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|---|---|
| CD | Course design, CD | |||||||
| 1 | Mini-lectures | 3.76 | 0.86 | 0 | 2 | 13 | 14 | 8 |
| 2 | Group activities | 3.05 | 0.88 | 1 | 7 | 21 | 5 | 3 |
| 3 | ImageJ | 3.08 | 0.92 | 0 | 11 | 15 | 8 | 3 |
| 4 | Printed 3D models | 3.57 | 0.80 | 0 | 3 | 14 | 16 | 4 |
| 5 | Virtual 3D models | 3.95 | 0.74 | 0 | 1 | 8 | 20 | 8 |
| 6 | Topics covered | 3.84 | 0.76 | 0 | 1 | 11 | 18 | 7 |
| 7 | Overall course | 3.70 | 0.74 | 0 | 1 | 14 | 17 | 5 |
| 8 | Course emphasis that imaging provides a way of knowing biology at different scales | 4.19 | 0.70 | 0 | 0 | 6 | 8 | 13 |
| 9 | NIH Roadmap | 3.54 | 0.93 | 0 | 5 | 13 | 13 | 6 |
| SG | Student gains, SG | |||||||
| 1 | Nanoimaging | 3.70 | 0.94 | 0 | 4 | 11 | 14 | 8 |
| 2 | Molecular imaging | 3.73 | 0.84 | 0 | 3 | 10 | 18 | 6 |
| 3 | System level imaging | 3.49 | 0.84 | 0 | 4 | 15 | 14 | 4 |
| 4 | Imaging as a quantitative tool | 3.76 | 0.86 | 0 | 2 | 13 | 14 | 8 |
| 5 | Biological scale | 3.76 | 0.86 | 0 | 3 | 10 | 17 | 7 |
| 6 | Biological imaging tools | 3.73 | 0.77 | 0 | 0 | 17 | 13 | 7 |
| 7 | Using imaging software | 3.73 | 0.80 | 0 | 1 | 15 | 14 | 7 |
| 8 | Interpreting imaging studies | 3.59 | 0.69 | 0 | 2 | 13 | 20 | 2 |
| 9 | Recognizing biological scale in images | 3.70 | 0.81 | 0 | 2 | 13 | 16 | 6 |
| 10 | Quantifying images | 3.46 | 0.84 | 0 | 3 | 19 | 10 | 5 |
| 11 | Importance of this field | 3.92 | 0.76 | 0 | 0 | 12 | 16 | 9 |
| 12 | Understanding that imaging offers new insights into biological structure and function | 3.86 | 0.86 | 0 | 1 | 13 | 13 | 10 |
| 13 | Understanding that imaging impacts medicine | 4.11 | 0.84 | 0 | 1 | 8 | 14 | 14 |
| 14 | Understanding that future patients will benefit from ongoing imaging research | 4.08 | 0.89 | 0 | 1 | 10 | 11 | 15 |
| 15 | Imaging is an integrated, multidisciplinary field | 3.70 | 0.88 | 0 | 3 | 12 | 15 | 7 |
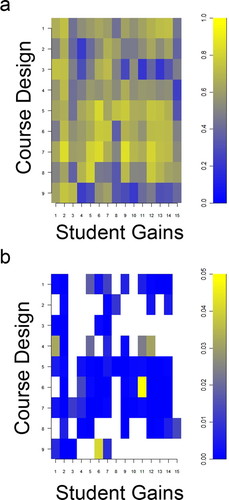
Figure 4. SALG association of course design with learning gains. (a) Polychoric correlations of course design with student gains in Table 3 were positive and indicated a positive alignment between active-learning activities and learning gains. (b) P values of significance (p ≤ 0. 05, corrected for multiple comparisons) for polychoric correlations in Figure 4A. Among other positive associations, the NIH Roadmap context in our course design contributed to positive gains in understanding nanoimaging (r = 0.61; p = 0.01; n = 37), molecular imaging (r = 0.75; p = 0.00; n = 37), and system level imaging (r = 0.71; p = 0.00; n = 37).
DISCUSSION
Our teaching unit used the NIH Roadmap as the central feature of course context and spanned a wide spectrum of imaging scales from nanoimaging to whole organisms. We used a scientific teaching approach to align learning goals with assessments and activities (Table 1). In this unit, we measured student learning through (1) pre- and postassessment of gained knowledge in biological scale, in a variety of imaging tools, and in the interpretation of biological images; (2) assessment activities drawn from the NIH Roadmap initiative; (3) the Gillespie rating scale to quantify student associations of biological utility with the quality of visual information contained in images; and (4) the SALG instrument, which allowed us to quantify associations between course design and student learning.
Summative measures of student learning gains indicated that our learning goals were achieved. Based on pre- and post- assessment, students made significant gains in understanding biological scale, distinguishing among a variety of imaging modalities, and interpreting images (Figure 1). These results respectively aligned with our learning goals, which were for students to understand the continuity of biological and imaging scale, the quantitative utility of imaging across a wide range of biological scales, and the importance of images for interpretation of structure and function.
In-class metrics indicated that our activities contributed to student learning gains. Students demonstrated proficiency with quantifying images using NIH ImageJ during the NanoBucky exercise (Figure 2). Incorporation of ImageJ into the course design contributed positively to gains in using imaging software (r = 0.71; p = 0.00; n = 37) and to various aspects of student learning (Figure 4). Using the Gillespie rating scale, students evaluated and perceived a strong association between the quality of visual information contained in biological images and the utility of these images within model types and an inverse association across model types (Figure 3, Table 2). The SALG instrument identified a strong association between the overall course and student gains in our learning goals including recognizing biological scale (r = 0.84; p = 0.00; n = 37) and understanding structure and function from images (r = 0.77; p = 0.00; n = 37). Furthermore, the NIH Roadmap context in our course design was associated with gains in nanoimaging (r = 0.61; p = 0.01; n = 37), molecular imaging (r = 0.75; p = 0.00; n = 37), system level imaging (r = 0.71; p = 0.00; n = 37), biological imaging tools (r = 0.56; p = 0.04; n = 37), and using imaging software (r = 0.61; p = 0.01; n = 37). Interestingly, student learning gains were not strongly associated with instructor evaluations (data not shown). This suggested that student learning was self-directed and took place within an active-learning framework as intended by our course design.
The current module would be useful as an introductory imaging unit in an introductory biology course. In addition, the NIH Roadmap initiatives we used to design this module provide a useful framework to introduce imaging concepts into the undergraduate, graduate, or medical school curriculum. Faculty with an imaging background would be best suited to teach an imaging module; however, those without an imaging background may want to separate the nanoscale, molecular, and medical imaging subunits and build upon the subunit most applicable to their overall course. The module in its current form could be adapted to a classroom of any size provided that computers and Internet access were made available. Students were provided with an overview of how the imaging equipment actually works and collects data. For example, with GFP, students were given a historical and practical overview of GFP discovery, the physics of fluorescence, and the meaning of excitation and emission spectra. Because this was an introductory course, we provided an overview of imaging quantification using images previously acquired with, for example, scanning electron microscopy and fluorescent imaging microscopy. Future implementations of this module as a full length course should incorporate finer details of image quantification by focusing on capture methodology including the importance of different exposures and neutral density filters for image correction. Future assessments using this course design may want to compare the utility of in-class activities to a purely lecture-based curriculum among two cohorts comprising those who attended lecture and those who attended lecture and participated in the active-learning activities to confirm the utility of the activities we developed based on the NIH Roadmap.
By using a scientific teaching approach and the NIH Roadmap to guide activities, this course made positive contributions to student gains in understanding that imaging is a useful way of knowing biology. This conclusion was based on summative pre-post assessments and in-class assessment using activities, Gillespie associations, and SALG associations. Early exposure to an imaging teaching unit addressed several misconceptions and may generate interest in advanced imaging education. Both the multidisciplinary nature of imaging and the NIH Roadmap context make this teaching unit accessible not only to students of biology but also physiology, engineering, mathematics, and physics. This teaching unit was limited by the number of class periods in the WOK course but may serve as a useful framework on which to build a longer, more in-depth course.
Accessing Materials
Materials for this imaging unit (including the syllabus, slides, pre/post questionnaires, Gillespie survey, SALG instrument, and imaging Internet resources) are freely available through the Wisconsin Program for Scientific Teaching (WPST) Digital Library website (http://scientificteaching.wisc.edu/materials/molecularbiology.htm).
ACKNOWLEDGMENTS
The Wisconsin Program for Scientific Teaching is supported by the Howard Hughes Medical Institute Professors Program awarded to Dr. Jo Handelsman, and by a grant from the National Science Foundation for Course, Curriculum, and Lab Improvement awarded to J.H. and Sarah Miller. This work was exempt from review with IRB protocol #2006-0516.



