Effectiveness of a Cloning and Sequencing Exercise on Student Learning with Subsequent Publication in the National Center for Biotechnology Information GenBank
Abstract
With rapid advances in biotechnology and molecular biology, instructors are challenged to not only provide undergraduate students with hands-on experiences in these disciplines but also to engage them in the “real-world” scientific process. Two common topics covered in biotechnology or molecular biology courses are gene-cloning and bioinformatics, but to provide students with a continuous laboratory-based research experience in these techniques is difficult. To meet these challenges, we have partnered with Bio-Rad Laboratories in the development of the “Cloning and Sequencing Explorer Series,” which combines wet-lab experiences (e.g., DNA extraction, polymerase chain reaction, ligation, transformation, and restriction digestion) with bioinformatics analysis (e.g., evaluation of DNA sequence quality, sequence editing, Basic Local Alignment Search Tool searches, contig construction, intron identification, and six-frame translation) to produce a sequence publishable in the National Center for Biotechnology Information GenBank. This 6- to 8-wk project-based exercise focuses on a pivotal gene of glycolysis (glyceraldehyde-3-phosphate dehydrogenase), in which students isolate, sequence, and characterize the gene from a plant species or cultivar not yet published in GenBank. Student achievement was evaluated using pre-, mid-, and final-test assessments, as well as with a survey to assess student perceptions. Student confidence with basic laboratory techniques and knowledge of bioinformatics tools were significantly increased upon completion of this hands-on exercise.
INTRODUCTION
GenBank is a comprehensive public database of annotated nucleotide sequences maintained by the National Center for Biotechnology Information (NCBI; Bethesda, MD). More than 103 million individual DNA/RNA sequences from >300,000 genera are currently available in this free, online database (Benson et al., 2009; NCBI, 2009). In 2005, Ostell reported that the NCBI website (www.ncbi.nlm.nih.gov) was visited by researchers throughout the world 50 million times per day, including 400,000 different homology searches.
The explosive growth in the number of nucleic acid sequences published in GenBank has made it the centerpiece for researchers worldwide. Reasons for accessing the GenBank database are as varied as the information stored there (Ostell, 2005; Sayers et al., 2009). Researchers use it to learn more about the identity, homology, structure, and variability of specific genes and gene products, as well as to explore gene function, gene expression, genome organization, and evolution.
In the past, many of the accessions published in GenBank were sequences from model organisms, such as yeast, fruit fly, mouse, and the plant Arabidopsis. Now that most model species have had their genomes sequenced, there is growing interest in examining the genes of lesser-studied organisms as well (Blaxter, 2002). To address the need for more data from nonmodel species, we developed a 6- to 8-wk laboratory exercise geared toward undergraduates that entails the cloning, sequencing, and bioinformatics analysis of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from a variety of plants.
The GAPDH enzyme catalyzes an important step in glycolysis and thus it occurs in all living organisms (Figge et al., 1999). As one of the most abundant enzymes in cells, GAPDH has served as a model protein for biochemists studying structure–function relationships, enzyme action, and metabolic pathways (Sirover, 1999). In humans, the GAPDH gene is highly expressed in 21 different types of cancer and may be associated with neuronal diseases such as Alzheimer's and Huntington's (Sirover, 1999; Altenberg and Greulich, 2004; Kim and Dang, 2005). The enzyme is also suspected to be involved in DNA replication and repair, cytoskeletal organization, and phosphotransferase activity (Tatton et al., 2000). Furthermore, GAPDH has been used to explore phylogenetic relationships in taxa as diverse as bacteria, crayfish, birds, and mosses (Figge et al., 1999; Wall, 2005; Ohlson et al., 2007; Mathews et al., 2008). Instructors, therefore, have opportunities to make connections between the molecular aspects of GAPDH and its biomedical and evolutionary significance.
Our students have been isolating and characterizing the GAPDH gene from numerous plant species since 2005. None of these sequences had been published previously in the NCBI GenBank. In the early years, we depended on more than a dozen individual commercial kits or products to carry out this multiweek exercise. Because our campus does not perform DNA sequencing, we contracted that service out to another university at $8 per run. To cover a significant portion of the GAPDH gene we designed three separate pairs of polymerase chain reaction (PCR) primers that annealed to three different regions, thus making it difficult to build a longer contig from the shorter DNA fragments. Although these challenges were not difficult to overcome, the project's complexity and expense might have discouraged potential instructors at other institutions. Although our goal was to widely disseminate this exercise and involve as many students as possible, we felt that a more streamlined process was needed. To make the GAPDH project more accessible to other institutions, we launched a collaborative effort with Bio-Rad Laboratories (Hercules, CA) and codeveloped an entire cloning and bioinformatics exercise. The resulting project is currently being marketed as the “Cloning and Sequencing Explorer Series” by Bio-Rad Laboratories. This novel exercise exposes students to a range of realistic research experiences, from the incorporation of numerous positive and negative controls to the discovery of GAPDH sequences from uncharacterized species. Likewise, the instructor benefits from lower costs, relative ease of preparation, use of fewer toxic reagents, and experimental robustness.
The GAPDH cloning exercise uses plants because of their high level of interspecies diversity, with >400,000 species on earth (Govaerts, 2001), as well as high intraspecies variability. For example, there are hundreds of cultivars of lettuce that could be studied (Ryder and McCreight, 2005). Furthermore, plant material for DNA extraction is readily available during all seasons from local gardens, plant nurseries, florists, and food stores. Another benefit is that instructors do not have to seek approval from their institutional animal care and use committees to extract DNA from plants.
The final product of this exercise is a unique and reliable DNA sequence for a GAPDH gene that can be published in the NCBI GenBank (Benson et al., 2009). These sequences are of great value to geneticists, biochemists, and evolutionary biologists worldwide. Because both the instructor and students are involved in obtaining and annotating the sequence they can be listed as coauthors on the GenBank accession. Furthermore, as the GAPDH gene database grows, it will become an even more valuable resource for instructors. At the same time, students benefit because they would feel part of a larger collaborative effort, and gain additional bioinformatics experience using these sequences to study plant phylogenetics.
The GAPDH cloning exercise incorporates important molecular laboratory techniques, as well as bioinformatics topics in a multifaceted approach to student learning. The student becomes involved in an investigative research project by using both a project-based exercise and active-learning strategy as recommended by the National Research Council (2003). These strategies teach basic laboratory methodologies (Handelsman et al., 2004), problem-solving skills, critical thinking, data analysis, and cooperative-group work (Regassa and Morrison-Shetlar, 2007). Student engagement in active-learning processes has been shown to improve knowledge retention (National Science Foundation, 1996; Handelsman et al., 2004; Halme et al., 2006). This exercise also addresses the recommendation of the American Society for Biochemistry and Molecular Biology that students become more familiar with bioinformatics tools (National Research Council, 2003; Boyle 2004). Bioinformatics (defined as the use of computers to collect, assemble, and analyze biological information) is gaining importance to biologists in all disciplines (Fuchs, 2002). The inclusion of bioinformatics in undergraduate textbooks and curricula demonstrates its relevance (Honts, 2003), as does the rapid growth of advanced degree programs in bioinformatics (Stein, 2008).
Our goals for the students in this laboratory-based project were as follows:
Students will gain competence in basic laboratory techniques in the context of a single research project.
Students will become familiar with a widely used bioinformatics software system (iFinch; Geospiza, Seattle, WA) to manage DNA sequence data, examine sequence quality, and resolve sequence ambiguity.
Students will develop skills using other bioinformatics tools, such as building contigs and predicting six-frame translation products.
Students will become comfortable using the NCBI website to perform and interpret various types of Basic Local Alignment Search Tool (BLAST) searches, to distinguish between different GAPDH loci, and identify intron/exon boundaries.
Students will produce a unique DNA sequence for GAPDH that may be published in GenBank with both instructor and student as coauthor.
To meet these goals, our students collectively cloned the GAPDH genes from four different plant species: coffee (Coffea arabica L.), purple heart (Setcreasea pallida Rose), sugarcane (Saccharum officinarum L.), and umbrella plant (Cyperus alternifolius L.). It was determined ahead of time that no GAPDH gene from these four species had been published in GenBank.
MATERIALS AND METHODS
In spring 2009, 27 students at Bellarmine University (Louisville, KY) were enrolled in a molecular biology lecture and laboratory course designed for junior-level science majors. Lectures were given three times per week, and students were divided into two 3-h laboratory sections that met once per week. Students worked in pairs. This exercise was initiated in the fourth week of the semester and continued for 7 wk (Table 1). Before its implementation, students were reintroduced to basic lab skills like micropipetting, gel electrophoresis, and PCR through a preliminary wet-lab exercise using PCR.
Week 1
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The Cloning and Sequencing Explorer Series (catalog no. 166-5000EDU), an eight-module kit, was purchased from Bio-Rad Laboratories. This kit contains all of the supplies and reagents, as well as directions for both the instructor and the students. The kit provides free DNA sequencing of 24 clones, carried out by the Department of Energy Joint Genomics Institute (DOE-JGI; Walnut Creek, CA). Included is contact information for obtaining DOE-JGI services and instructions for setting up the free, individual classroom account for accessing iFinch bioinformatics software.
At the beginning of the exercise, students were provided with a comprehensive lab manual that included relevant background information, lab instructions, worksheets, and detailed bioinformatics software directions. This lab manual, written in collaboration between us, Bio-Rad Laboratories, and Geospiza, is provided with the kit. Students were expected to read each week's assignment before attending lab. Labs were generally preceded by short lectures on relevant topics such as DNA extraction, PCR-primer design, ligation, transformation, gel electrophoresis, the Sanger method of DNA sequencing (Sanger and Coulson, 1975), contig building, and BLAST searching.
Cloning and Sequencing the GAPDH Gene
DNA Isolation. Each group of two students was assigned one of the plant species to work with, and each species was assigned to at least three groups. Students collected juvenile leaves from plants (purchased from horticultural suppliers) that were growing in the campus greenhouse. Leaf tissue was placed into a microcentrifuge tube and ground with a micropestle in standard lysis buffer at room temperature. Genomic DNA was then extracted using silica-based chromatography columns according to the manufacturer's protocol. To prevent contamination, filter barrier tips were used in the DNA extraction step, and in all subsequent steps.
Amplification of the GAPDH Gene. The kit takes a nested PCR approach involving two separate amplification steps (Dieffenbach and Dveksler, 2003). For organisms whose genomes are not fully characterized, the nested PCR approach allows for greater specificity because four primers must anneal to a single region of the target gene rather than just two primers. The first PCR amplification step used degenerate primers based on a consensus sequence of plant GAPDH. Of the seven known loci in Arabidopsis, two encode enzymes that are cytosolic and the remaining five are localized to plastids. The nested primers were designed to amplify the cytosolic forms, GAPC and/or GAPC-2. Approximately two-thirds of the gene is targeted in this project, which is equivalent to 60% of the GAPC coding region in Arabidopsis, and includes the enzymatic active site. Students set up initial PCR reactions in 40-μl volumes by using 5 μl of DNA template, and diluted the initial primers to a final concentration of 2 μM according to kit instructions. Two positive controls (Arabidopsis genomic DNA and a plasmid-borne Arabidopsis GAPDH clone) and one negative control (water) were also prepared by each group in both rounds of PCR. DNA quantification was not required for PCR amplification, but it is an option for instructors.
After amplification, unincorporated primers were removed by treatment with exonuclease I, which was then heat inactivated. The initial amplification product served as the template for the nested PCR reaction, which also used consensus primers that were designed internal to the initial PCR primers. Nested PCR reactions were performed with 20 μl of 100× diluted template in a 40-μl volume according to instructions. Agarose gel electrophoresis (1%) was performed and then visualized with ethidium bromide to confirm PCR success.
PCR Purification, Blunt-End Digestion, Ligation, and Transformation. Size-exclusion chromatography columns were used to purify the nested PCR product. To produce “blunt-ends” for ligation, the 3′-adenosine nucleotide was removed using a proofreading polymerase. Samples were incubated at 70°C for 5 min and then cooled on ice for 2 min. Ligation to blunted pJet 1.2 parental vector was performed at room temperature for 10 min. Students made Escherichia coli (HB101) cells competent, and heat-shock transformation was performed on ampicillin selection plates containing isopropyl β-d-1-thiogalactopyranoside prewarmed to 37°C. Selection for transformants was also made possible by the disruption of the eco47IR lethal gene in the pJet 1.2 vector. A positive control (transformation with a plasmid-borne Arabidopsis GAPDH clone) was also carried out by each student group.
Plasmid Preparation and Restriction Enzyme Digestion. Before the fourth lab period, each group picked four putatively transformed colonies using sterile toothpicks and grew them overnight with shaking at 37°C in liquid Luria-Bertani (LB) growth media containing ampicillin. Plasmid minipreps were performed using the Aurum Plasmid Mini Kit (Bio-Rad Laboratories) included with this exercise. Mini-prepped DNA was digested with BgIII restriction enzyme, which cuts on both sides of the insert. Gel electrophoresis (1% agarose) was performed by students to confirm the presence of an insert and determine its size.
DNA Sequencing. Quantification of DNA was not required for DNA sequencing, but is an option. Each group chose one or two clones containing the expected insert size to send to DOE-JGI for sequencing. For each clone, 10 μl of mini-prepped plasmid was mixed separately with four different sequencing primers to ensure full-length coverage of the insert on both strands. Two of the sequencing primers were designed to recognize the pJet vector (flanking both sides of the insert). The other two sequencing primers were based on internal conserved regions within GAPDH exons. One vector primer and one internal primer were used to attain full coverage of each strand. Twenty-four clones were fully sequenced using a 96-well plate mailed to DOE-JGI. However, students can perform their own DNA sequencing if the equipment is available.
Bioinformatics Analysis of the GAPDH Gene
Sequencing results were posted on a secure website within 2 wk. During the interim, students were introduced to the iFinch software system using practice data published at http://www.geospiza.com/education/products/Bio-Rad.html, which is described in Supplemental Material 1. In the pre-exercise, students examined the quality of a sequencing read, assembled a contiguous DNA sequence from four shorter overlapping sequences, performed a BLASTn search using the generated contig to identify homologous sequences, and examined phylogenetic relationships. Students collaborated in groups of two on all bioinformatics exercises, but all assignments were evaluated individually.
Table 2 provides an overview of the bioinformatics analysis that was performed on the student's clones. A secure iFinch account was established as described in the instructor's manual. iFinch software contains links to other relevant bioinformatics tools. Instructors organized the 96 DNA sequences into folders for each group, with each folder containing four sequence files per clone. Low-quality reads were automatically identified by iFinch, as was the pJet 1.2 vector sequence. Through an automated process, iFinch can identify and count the number of clones that match GAPDH. The software allows the instructor to estimate which GAPDH genes have been cloned and how many clones have been obtained, immediately after uploading student data. Students examined traces to determine the quality of the reads with the FinchTV chromatogram viewer (Geospiza) and used BLASTn to obtain a preliminary identification. The four sequences from each clone were then assembled into a larger contiguous sequence using a program linked to the iFinch site. Base pair discrepancies between overlapping sequences were identified and chromatograms were examined in detail using FinchTV software. Where appropriate, students edited bases and reassembled the contig.
| 1 | Examine quality of DNA sequence traces using FinchTV |
| 2 | Evaluate the components that make up the quality scores |
| 3 | Distinguish vector from GAPDH sequence |
| 4 | Convert sequence data to FASTA format |
| 5 | Batch BLASTn search against plant ″reference genomic sequences″ |
| 6 | Interpret BLAST results |
| 7 | Assemble contig from the four individual reads |
| 8 | Edit discrepancies between the four reads |
| 9 | Reassemble contig from edited reads |
| 10 | Identify which GAPDH gene was cloned using BLASTn of contig against plant ″reference genomic sequences″ |
| 11 | Determine intron/exon structure using BLASTn against plant ″reference mRNA sequences″ |
| 12 | Locate exact intron/exon boundaries and construct a predicted mRNA sequence |
| 13 | Confirm predicted mRNA sequence using a BLASTn search against plant ″reference mRNA sequences″ |
| 14 | Determine the amino acid sequence of the predicted mRNA using BLASTx against plant ″nonredundant protein sequences″ |
| 15 | Six-frame translation of the predicted mRNA |
| 16 | Identify the correct reading frame for the predicted mRNA |
| 17 | Finalize a publishable contig sequence for each plant species by comparing the different clones |
Seven different GAPDH loci have been identified in the plant model species Arabidopsis thaliana. Students identified which GAPDH gene they most likely cloned based on their BLASTn alignment scores. Exon/intron boundaries were identified by looking at genomic and mRNA sequences from different plant species. Students removed potential intronic regions, performed a six-frame translation to obtain a predicted amino acid sequence, and then used BLASTp to verify their mRNA sequence. Afterward, each group's work was confirmed by comparing the different contigs from a single plant species to each other.
Evaluation of Student Outcomes
Learning Assessment. A pretest consisting of 12 multiple-choice questions was given to all students the first day of class (Supplemental Material 2, questions 1–12). Students were also asked to self-report their confidence level about their answers using a 5-point scale, where 1 is “lowest confidence” and 5 is “highest confidence.” One of the optional answers for each multiple-choice question was “I have no idea,” which was automatically scored as lowest confidence. The same test was given two more times during the semester: during week 5 (midtest), after their clones had been mailed for sequencing, but before the introduction of iFinch, and again at the end of the semester (final test). An additional four questions (13–16) about bioinformatics were asked on the midtest, and the same questions were asked on the final test. To reduce bias, tests were never returned and specific answers to questions were not disclosed. Overall test scores were compared using a one-way analysis of variance (Jandel Scientific, 1995). Statistical differences between individual pre-, mid-, and final-test questions were determined by the Wilcoxon signed rank test. Also, correlation analysis was performed on self-reported confidence levels versus student performance on test questions (SPSS, 2002).
Student Perception Survey. Three weeks after completing the GAPDH cloning and bioinformatics exercises, students were given an anonymous 12-question survey (Table 3) to assess student perceptions about their achievement of the five learning goals. The survey used a 5-point Likert scale: “strongly disagree,” “disagree,” “neither disagree nor agree,” “agree,” and “strongly agree” (Likert, 1932). Questions 1 versus 2 and 3 versus 4, which related to student comfort levels on wet-lab and bioinformatics skills, were compared using the Wilcoxon signed rank test (Jandel Scientific, 1995). Students were also asked to anonymously write their personal opinions about the exercise.
| Question | Avg. | SD |
|---|---|---|
| 1. Before the semester began, I was very comfortable with basic laboratory techniques, such as pipetting, centrifuging, and loading gels. | 4.19 | 0.801 |
| 2. Overall, the wet-lab exercises improved my comfort level with basic laboratory skills, such as pipetting, centrifuging, and loading gels. | 4.73 | 0.452 |
| 3. Before the semester began, I was familiar with the basic bioinformatics tools for examining DNA sequences, building DNA contigs, performing BLAST searches, and identifying introns. | 2.85 | 1.084 |
| 4. Overall, the bioinformatics exercises helped me become more familiar with basic skills, such as examining DNA sequence, building DNA contigs, BLAST searching, and identifying introns. | 4.46 | 0.706 |
| 5. Overall, this lab improved my understanding and appreciation for the processes and techniques used to clone and analyze genes. | 4.54 | 0.508 |
| 6. I have a better understanding of the process of cloning and bioinformatics after performing those exercises than if I had just heard about them in lecture or read a textbook. | 4.65 | 0.485 |
| 7. Cloning the GAPDH gene was easier than I initially thought it would be. | 3.35 | 0.940 |
| 8. Bioinformatics was easier than I initially thought it would be. | 3.46 | 1.030 |
| 9. I feel more comfortable using the NCBI website now than I did before starting this exercise. | 4.42 | 0.703 |
| 10. This lab gave me a better appreciation of bioinformatics and how computers are used to analyze DNA sequence. | 4.50 | 0.583 |
| 11. I would recommend having students carry out these exercises in this course in the future. | 4.31 | 0.679 |
| 12. I would be interested in seeing my name published in an NCBI GenBank accession. | 4.65 | 0.629 |
Technical Abilities. Student technical abilities were monitored by the instructor at key points throughout the process. Successful amplification of the two positive controls, along with lack of amplification of the negative water control were indicators of student technical ability, as were transformation efficiency, plasmid purification, and restriction digest results.
Student comprehension of bioinformatics also was assessed. During the laboratory sessions the instructor continually examined students' results after BLAST searches, contig assembly, intron/exon identification, and six-frame translation. Lab books and printouts of assembled contigs, intron predictions, and sequence alignments were completed by students and collected at the end of the exercise.
RESULTS
Cloning and Sequencing the GAPDH Gene
DNA Isolation. Based on the PCR results described in the next section, genomic DNA seemed to have been successfully isolated from coffee, purple heart, sugarcane, and umbrella plant leaves.
Amplification of the GAPDH Gene. The plasmid positive-control was amplified by students in both initial and nested PCR rounds with 100% success. The Arabidopsis genomic positive control was successfully amplified by 85% of the groups in the initial PCR step and by 92% of the groups in the nested PCR step. An example of an unsuccessful amplification of the Arabidopsis genomic control after a nested PCR reaction is shown in Figure 1A, lane An, and was most likely due to pipetting error or insufficient mixing (the most common technical error). One of the negative controls (water) showed an amplification product after the nested PCR step (data not shown), probably due to a student using a contaminated pipetter tip or misidentifying a PCR tube.
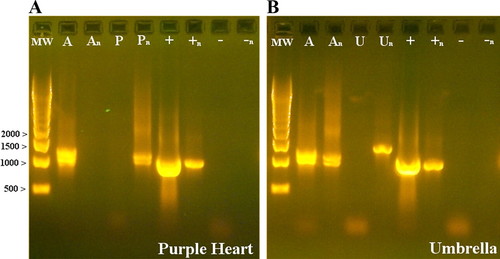
Figure 1. Gel electrophoresis results after initial and nested PCR reactions for two plant species. A 500-bp molecular weight (MW) ladder was used for size estimation. Initial PCR of the two positive controls: genomic DNA from Arabidopsis (lane A) and plasmid DNA of the cloned GAPDH gene from Arabidopsis (lane +). Nested PCR of the same two positive controls are shown in lanes An and +n. The band in lane +n seems larger than the band in lane + because of unequal loading (5 vs. 20 μl of sample, respectively). Water served as the negative control (lane −). Initial PCR of genomic DNA extracted from purple heart (lane P) is shown in Gel A, whereas DNA from umbrella plant (lane U) is shown in Gel B. Initial PCR product served as the template for the nested PCR reactions (lanes An, +n, −n, Pn, and Un). The multiple bands observed with the Arabidopsis control (lanes A and An) result from consensus primers annealing to more than one GAPDH locus.
All 13 groups observed amplification products for their plant samples using the GAPDH primers. The three groups of students working with sugarcane saw an amplification product after both the initial and the nested PCR steps (data not shown). Students working with purple heart and umbrella plant consistently observed PCR products after the nested step but not after the initial PCR reaction (Figure 1A, lanes P vs. Pn, and B, lanes U vs. Un). This was to be expected, because the initial PCR was performed using degenerate, consensus primers that were designed to be robust but can result in lower amounts of initial PCR product. It is not uncommon for initial PCR products to be invisible on the gel when using genomic DNA from distantly related species. In this scenario, there is usually enough amplicon to serve as a template for the nested round of PCR. Two of the four groups working with coffee DNA observed both the initial and nested PCR product, whereas the other two groups observed nested PCR products only (data not shown). These inconsistencies could be due to differences in the concentration of DNA template used for the initial PCR.
PCR Purification, Blunt-End Digestion, Ligation, and Transformation. Because every group obtained nested PCR products from their respective plants all of the students were able to proceed with the PCR purification step. Once purified, the PCR products were blunt-ended, ligated to vector, and used to transform chemically competent E. coli. Success was monitored by the number of groups obtaining colonies capable of surviving on selection plates. Twelve of 13 groups (92%) succeeded at ligation and transformation. Typically, each plate contained >100 colonies growing on them.
Plasmid Preparation and Restriction Enzyme Digestion. Each group picked four colonies from their plate by using a sterile toothpick and grew them overnight in LB growth media broth containing ampicillin. The group without their own transformants plucked colonies from another group's plate that contained clones from the same purple heart plant. Plasmid minipreps were performed, and plasmids were restriction enzyme digested to confirm inserts via gel electrophoresis (Figure 2). One undigested clone from each group was included in the gel for comparison. In gel A (Figure 2), several bands can be seen in the undigested lane compared with gel B (lane 11u vs. 51u). The most prominent band is probably supercoiled plasmid DNA, whereas the upper bands could be sheared or nicked plasmid that migrates at higher molecular weights. Nicking and shearing of plasmid DNA are artifacts of plasmid purification but also could indicate rough handling of the sample. This scenario offers another opportunity to discuss key principles of gel electrophoresis, DNA structure, and the importance of student technique.
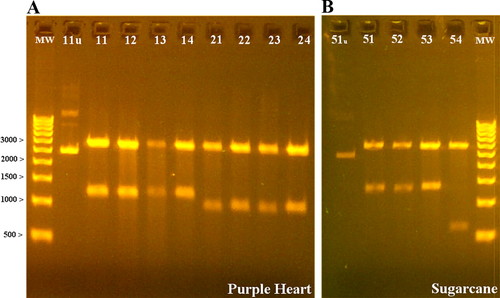
Figure 2. Gel electrophoresis results after BglII restriction enzyme digestion of putative GAPDH clones. A 500-base pair molecular weight ladder (MW) was used for size estimation. Gel A represents eight clones from purple heart (lanes 11–14 and 21–24). An undigested sample (lane 11u) was used for comparison. Gel B represents four putative GAPDH clones from sugarcane (lanes 51–54), as well as an undigested sample (lane 51u). The cloning vector used was approximately 3000 base pairs.
Of the 54 colonies examined by gel electrophoresis, 35 contained inserts of the expected size. It seemed that most of the negative results were due to technical problems during plasmid purification or restriction digest. One group, for example, broke the membranes of four spin columns with a pipette tip. An additional three clones contained inserts that were deemed too small to be GAPDH and thus were not considered. Two clones, both from a single group, seemed to contain degraded DNA, even after repeating the restriction enzyme digest. DNA degradation probably occurred during plasmid purification possibly due to nuclease contamination. The remaining samples seemed to be either undigested plasmids or did not contain insert.
Figure 2A shows two distinct insert sizes of approximately 1.2 kb (lanes 11–14) and 1.0 kb (lanes 21–24). Two different insert sizes were expected because the nested PCR for purple heart produced two bands (Figure 1A, lane Pn). These groups all used the same purple heart plant as their source of genomic DNA; therefore, the two sizes probably represent two different loci. Figure 2B also shows two different insert sizes, one insert at approximately 1.3 kb (lanes 51–53) and the other insert at approximately 0.6 kb (in lane 54). There are several plausible explanations for the 0.6-kb band, including cloning of fragments such as short PCR extensions (common to nested PCR) or degraded PCR products. To elucidate the identity of the smaller insert, the clone would need to be sequenced.
DNA Sequencing. Because a single 96-well plate is used for DNA sequencing, and each clone requires four different wells, only 23 of the 35 putative clones (plus one plasmid-borne Arabidopsis GAPDH control) could be sequenced. Each group sent out at least one of their clones for sequencing, and most sent out two. DNA sequence data were received from DOE-JGI within 2 wk.
Bioinformatics Analysis of the GAPDH Gene. Students successfully performed all steps outlined in Table 2. Of the 23 clones sent to DOE-JGI for sequencing, 16 (70%) were identified as the GAPDH gene, with the remaining seven clones either containing nontarget insert DNA or low-quality reads (defined as a read with <50 bases that have quality scores >20). Some low-quality reads are to be expected with large numbers of sequencing reactions. Of the eight purple heart clones submitted for sequencing, all were homologous to either the GAPC or GAPC-2 loci (for cytosolic GAPDH) in Arabidopsis. Five clones each from sugarcane and umbrella plant were submitted for sequencing, with three from each species being similar to plant GAPC. Of the five coffee clones submitted, two were homologous to plant GAPC.
Evaluation of Student Outcomes
Learning Assessment. On the first day of class, students were given a pretest consisting of 12 multiple-choice questions relevant to gene cloning and bioinformatics (Supplemental Material 2, questions 1–12). Students also were asked to self-report their confidence levels for their answers to each question. On average, students answered 46% of the questions correctly (Figure 3A), with a self-reported confidence level of 2.55 (of 5.00; Figure 3B). The two lowest scores (22%) were on questions 10 and 12 related to sequencing and bioinformatics (Figure 4). Self-reported confidence levels (Figure 5) were also lowest on these two questions, 1.48 and 1.44 (of 5.00), respectively. Highest pretest scores (>74%) were for questions 1, 4, and 7 that concerned the role of GAPDH in metabolism and the basics of gel electrophoresis, both topics that were covered in previous courses (Figure 4).
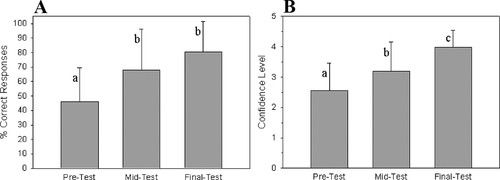
Figure 3. Overall averages (n = 27) for the pretest given at the beginning of the semester, the midtest given during week 5, and the final test given at the end of the semester. Graph A represents percentage of correct responses ± SD. Graph B represents self-reported confidence levels about their responses ± SD. One-way analysis of variance was used for comparison. Averages identified by a different letter are statistically different (p < 0.05).
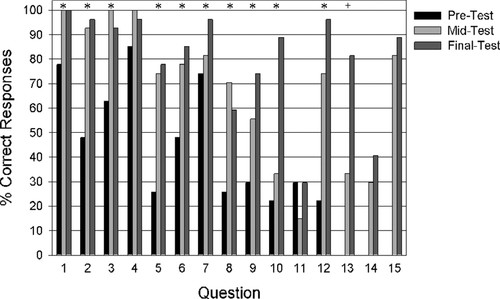
Figure 4. Percentage of correct responses (n = 27) for individual questions on the pretest given at the beginning of the semester, the midtest given during week 5, and on the final test given at the end of the semester. Asterisk (*) indicates significant differences (p < 0.05) from pretest to final test. + indicates significant differences (p < 0.001) from midtest to final test.
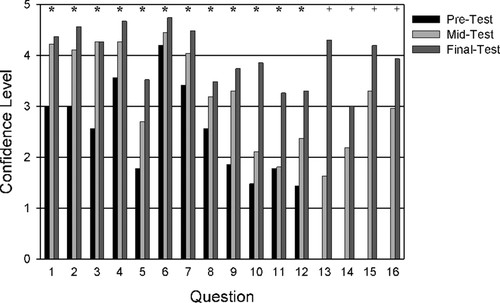
Figure 5. Self-reported confidence levels (n = 27) for individual questions on the pretest given at the beginning of the semester, the midtest given during week 5, and on the final test given at the end of the semester. Asterisk (*) indicates significant differences (p < 0.01) from pretest to final test. + indicates significant differences (p < 0.01) from midtest to final test.
One week after the conclusion of the wet-lab exercises, but before beginning the bioinformatics portion, students were given the same test, but with four additional bioinformatics- related questions (Supplemental Material 2, questions 13–16). Responses to the initial 12 questions on the midtest averaged 73%, which was an increase of 58% over the pretest (Figure 3A). The average student confidence level on the same 12 questions was 3.40, an increase of 33% (Figure 3B). Both of these increases were significantly different (p < 0.001). On the midtest, the lowest score (15%) was on question 11, which was about the Sanger method of DNA sequencing (Figure 4). Although question 11 was the only example of a midtest score being lower than the pretest score, this decrease was not statistically significant (p = 0.125). Of the three new questions about bioinformatics (13–15), 48% were answered correctly.
At the end of the semester, students were given the same test questions again. Students correctly responded to all 15 multiple-choice questions 80% of the time (Figure 3A), with a self-reported confidence level of 4.0 (Figure 3B). This represents an 18% increase in the number of students answering the questions correctly compared with the midtest (not statistically significant, p = 0.063). Of the 12 questions asked on both the pretest and final test, there was an 80% increase in correct responses (significantly different, p < 0.001).
Overall, student confidence levels increased throughout the semester (Figure 3B). At the beginning of the semester, students reported an overall confidence level of 2.55; by the midtest, confidence levels rose to 3.2; and at the final test, confidence levels were 4.0. The confidence levels on each of these tests were significantly greater than the previous (p < 0.001). There was a significant, positive correlation between the average test score for each question, and average self-confidence on that question (r2 = 0.71, p < 0.001). Self-reported comfort levels about managing DNA sequence files (Supplemental Material 2, question 16; and Figure 5) also increased significantly from the midtest to the final test (p < 0.001).
Students showed the greatest overall improvement for questions 5, 9, 10, 12, and 13 (Supplemental Material 2 and Figure 4). This indicates that students came into the class with less experience on topics such as bioinformatics, DNA sequencing, and cloning techniques. The most difficult question for students throughout the semester seemed to be question 11 (which no more than one-third of students answered correctly) regarding which region of the output is typically most reliable when using the Sanger method of DNA sequencing. No statistically significant differences (p > 0.05) were observed in the scores for question 11 on the pre-, mid-, and final test. Perhaps rephrasing question 11 to “When using the Sanger method to determine the sequence of a cloned gene, which region produces the most reliable output?” would help clarify the question. Question 14, which concerned the appropriate NCBI database and BLAST searches to use when analyzing mRNA sequences, also showed low levels of improvement, from 30% on the midtest to 41% on the final test (not significantly different, p = 0.25). No statistical improvement was observed for question 15, for which students were asked to apply BLAST criteria in comparing different DNA sequences, because scores were already fairly high on the midtest.
Student Perception Survey. At the end of the semester, the class was given an anonymous survey to assess their perceptions of their achievement of goals in the exercise (Table 3). Students reported that they felt fairly comfortable (4.19 of 5.00) with standard laboratory techniques such as pipetting, centrifuging, and gel electrophoresis at the beginning of the semester (question 1). As indicated by question 2, the experiences gained from this laboratory exercise increased their comfort levels with these techniques by 13% (significantly different, p = 0.017). Student comfort with bioinformatics tools, such as examining DNA sequences, building contigs, and intron identification was fairly low (2.85 of 5.00) at the start of the semester (question 3). Familiarity with bioinformatics skills (question 4) increased 56% by the end of the exercise (significantly different, p < 0.001). Even though students thought that they had learned a great deal about cloning and analyzing genes (questions 5 and 6), they still believed that the process was fairly challenging (questions 7 and 8). The majority (88%) of the class recommended this exercise for future years (answered agree or strongly agree for question 11). Likewise, 92% of the students reported that they were very interested in seeing their name published in the NCBI GenBank (answered agree or strongly agree for question 12).
Students also wrote general comments about their experience with the Bio-Rad Cloning and Sequencing Explorer Series. Students commented that they enjoyed the unique, in-depth, “more involved” laboratory experiences that were used to pursue a single research topic for numerous weeks. Most reported that they realized the importance of reading and following directions carefully, of accurate pipetting skills, and of maintaining good group dynamics. They also felt that the information and techniques used in this kit were well connected with the class lecture material and were relevant to real-world research. Furthermore, the class enjoyed the idea that they would generate data that was publishable and that could be helpful to scientists or even students working on GAPDH at other institutions. It was suggested that quizzes and homework assignments be incorporated into the laboratory as a way of encouraging them to keep up with the material.
Students also commented that the bioinformatics portion of the exercise was more challenging than the wet-lab portion. Students reported that they felt more comfortable using the NCBI website by the end of the exercise, as well as using the iFinch system. They wrote that they gained a better appreciation for the field of bioinformatics and enjoyed learning about the different BLAST programs, how to edit DNA, build contigs, and identify introns. Some students initially had problems with Web browser incompatibility when using Macintosh computers, but these issues can be overcome by switching to alternative Web browsers such as OmniWeb, Firefox, or Internet Explorer. Other institutions regularly use iFinch on Macintosh computers without problems.
Technical Abilities. Technical abilities were continually monitored by the instructor. Overall, the amplification steps for the GAPDH gene were highly successful, with 100% of the groups obtaining nested PCR product of the appropriate size. All groups were then able to purify their PCR products and prepare for ligation and transformation. One (8%) of the 13 groups was unsuccessful in obtaining transformants, however. That group also failed to obtain transformants of the positive plasmid-borne Arabidopsis GAPDH control, indicating that the problem was during the heat-shock transformation step.
Of the 54 putative clones that were miniprepped, 91% contained plasmid DNA. Student errors during the miniprep procedure accounted for the 9% that did not contain plasmid. Taking into account only the clones containing plasmids, 70% seemed to contain an insert of the appropriate size after restriction digest. Twenty percent of the clones containing plasmid did not produce an insert after digestion. These could be false positives, although the lethal eco47IR gene should have prevented bacterial cells without inserts from surviving. Another possibility is that short DNA molecules such as primer-dimers were cloned but not detected because they ran off the gel. Two of the clones that did not produce an insert were similar to the undigested control, which indicates students had problems with pipetting or that the enzyme was not thoroughly mixed with the plasmid. Two samples (4%) were so degraded that the plasmid was unrecognizable. Another 6% contained small inserts deemed to be PCR artifacts, such as nonspecific amplification product from the initial PCR round.
The GAPDH gene was successfully identified from 70% of the plasmid DNAs sent to DOE-JGI for sequencing. Because DNA concentrations were not quantified before sequencing, 70% can be considered successful. The targeted GAPDH sequence was obtained from 100% of the purple heart clones, 60% from both the sugarcane and umbrella plant clones, and 40% of coffee clones.
Student success at bioinformatic analysis was monitored by the instructor at various stages. Students were introduced to iFinch software system through a pre-exercise (Supplemental Material 1) during week 5. The average score on the pre-exercise was 81%. During weeks 6 and 7, tasks such as contig construction and sequencing edits were printed by each group and were turned in. The instructor examined and discussed BLAST searches, intron/exon identification, and six-frame translation with each group as results were generated. At the end of the semester, students turned in their answers to the numerous questions posed in the iFinch manual. The answers were evaluated by the instructor and the average score received was 87%.
DISCUSSION
The GAPDH cloning exercise introduces students to a multitude of wet-lab techniques, such as DNA isolation, PCR, gel electrophoresis, PCR purification, preparation of competent cells, ligation, heat-shock transformation, plasmid isolation, and restriction digests. The bioinformatics portion of the exercise allows students to follow through with the project and introduces them to important concepts, such as homology searching, contig construction, intron/exon identification, and sequence alignments, all of which go into annotating a publishable GenBank accession.
Overall, it seemed that students benefitted from the hands-on approach taken by the GAPDH cloning and bioinformatics exercise. Students reported greater understanding of the techniques used in cloning, increased competence in their laboratory ability, and a better appreciation of bioinformatics tools. The instructor also noticed that students were more confident and accurate in their pipetting, preparation of PCR reactions, and gel loading. Further evidence of their competence was reflected by the high rates of success in cloning the GAPDH gene. Faculty at our institution also commented that students who have completed the GAPDH cloning exercise seem more comfortable with basic molecular techniques when working on independent research projects. Another form of assessment was improved performance on the pre-, mid-, and final test, which covered a wide range of cloning and bioinformatics topics.
All five of our student-learning goals were successfully met by the GAPDH cloning and bioinformatics project. Our students had a 92% success rate in obtaining GAPDH clones, which compares favorably with the 70–80% success rate for a subcloning project by using a bioluminescence gene reported by Regassa and Morrison-Shetlar (2007). Therefore, goal 1 was met. Students reported that they became more familiar with managing DNA data files, as well as assessing sequence quality and resolving DNA base ambiguities. They also seemed to appreciate that they were taking part in an entire research experience, from the isolation of a gene to the publication of its sequence. During face-to-face conversations with groups during the bioinformatics portion of the exercise students seemed to recognize the potential, as well as limitations, of computer programs for assembling contigs, identifying intron/exon boundaries, and predicting protein sequences. For example, students discovered that a minimum number of overlapping bases are needed to build a contig from shorter sequences and that six-frame translation of DNA is difficult if the DNA sequence is inaccurate. Although students felt challenged using iFinch and other bioinformatics tools, they reported that they learned a great deal. Therefore, we feel that goals 2–4 also were successfully achieved.
Student enthusiasm for the GAPDH cloning project remained high throughout the semester, with the majority recommending the exercise be used in next year's class. Student interest also can be demonstrated by the large number (72%) who volunteered to come in during the summer to help annotate the final sequence for submission to the NCBI GenBank. In our experience, most follow through with these commitments. Currently, 22 (of 59) of our students have been listed as coauthors on GAPDH accessions already published in GenBank. Our students seem motivated by the desire to cite these accessions on their resumes. Our policy is to require extra effort from the student for coauthorship, but it is at the discretion of the instructor. One institution using the GAPDH cloning and bioinformatics kit has 18 coauthors listed in its GenBank accession. The ultimate publication of a GAPDH sequence demonstrates our success at meeting goal 5.
This exercise provides ample opportunities to stimulate critical thinking. Throughout the wet-lab portion of the exercise, students had to calculate PCR and ligation volumes, interpret gel-electrophoresis results, and participate in group decision-making about upcoming steps. During the bioinformatics portion of the exercise students had to make judgments about sequence ambiguity and make nucleotide edits when appropriate. They also had to analyze numerous BLAST results and discern which particular member of the plant GAPDH gene family their clone most closely resembled. In addition, students were expected to answer numerous critical-thinking questions in their lab-books. These questions concerned concepts such as positive and negative controls, restriction mapping of their clone, sequence-quality parameters, and gene structure.
The GAPDH cloning and sequencing exercise is highly transferable and easily adaptable to a wide range of undergraduate and early graduate-level courses, as well as to advanced level biotechnology courses in high school. It also could be included into the biotechnology curriculum at community or technical colleges. The exercise is flexible enough that it can be completed within a half to a full semester. Students only had to work outside of the scheduled lab period once: to select transformed colonies for subsequent plasmid isolation.
This exercise is both instructor and student friendly. Genomic DNA extraction did not require the use of liquid nitrogen or dry ice, and plasmid isolation did not involve hazardous chemicals such as phenol and chloroform. The two PCR steps seemed to be very robust, because the primers worked on a variety of plant species. Even though the exercise uses blunt-end ligation of the PCR product to the cloning vector, bacterial transformation efficiency was very high. Although quantification of genomic, amplified, or plasmid DNA is not required it could be integrated at one step or another.
We also appreciated the flexibility of the iFinch software. Given that iFinch is a web-based system, data stored in iFinch can be accessed wherever an Internet connection is available. Because iFinch tracks changes and preserves the original chromatograms, as well as the versions edited by students, the instructor does not have to worry about losing vital information.
For the 27 students enrolled in 2009, the final cost of our 7-wk exercise was $55/student. This compares favorably with Bramer et al. (2008) at $100/student for an entire semester-long project and with Hood-DeGrenier (2008) at $26/student for a 3-wk exercise. When we first initiated the GAPDH cloning exercise (in 2005 with 16 students), we spent approximately $115/student to carry out just the wet-lab portion. At that time, bioinformatics analysis was performed with VectorNTI software (Invitrogen, Carlsbad, CA), which was freely available but now costs $495 per computer. The Bio-Rad Cloning and Sequencing Explorer Series consists of eight sequential modules, each sold individually, and several do not require yearly purchases, thus reducing costs in future years.
The bioinformatics portion was reported by students to be more difficult than the wet-lab portion. Students generally scored lower on test questions covering bioinformatics (Supplemental Material 2, questions 11–15), indicating the challenges associated with that discipline. Most undergraduates are unfamiliar with database management and bioinformatics software systems, so a learning curve needs to be overcome for using systems such as iFinch. Students were sometimes overwhelmed with the length and complexity of bioinformatic instructions. However, we feel that familiarity with bioinformatics software is critical, because bioinformatics is becoming an important part of any biology profession (Stein, 2008).
Many students commented that this was their only opportunity to explore bioinformatics in depth. To increase their comfort level, students recommended homework assignments on the various bioinformatics tools. They also suggested that the narratives on bioinformatics that are provided with the kit be thoroughly read and understood before lab; so, in the future we plan on giving more prelab lectures and quizzes. Another option is to assign the tutorials and short videos that are freely available on the iFinch home page (described in Materials and Methods). These learning aids provide practical instructions on using iFinch, editing traces with FinchTV, building contigs, and BLAST searching. These tutorials and movies were not assigned in our class in 2009 but will be required next year.
We recommend that a minimum of three different clones from the same individual plant be sequenced in both the forward and reverse directions. This is why it is useful to have multiple groups of students researching the same plant species. Another advantage of having multiple groups studying the same species is that if one group is unsuccessful at any particular stage, a different group working with the same plant can share their material. For example, students have 15–30× more nested PCR product than they need for ligation and transformation. Likewise, as experienced this year, poor ligation or transformation efficiency by one group can easily be resolved by using another group's colonies from the same plant. We also had each group select and purify plasmid from four different putative transformants, thereby reducing the need for repeating minipreps or restriction digests in cases of experimental error. Because multiple clones were sequenced from the same plant, a poor read from one could be compensated by examining other clones. Due to this redundancy, our students did not have to repeat any experimental steps in 2009, although that is always an option.
In the future, after students have completed the GAPDH exercise, we are planning on having them construct a concept map outlining the individual steps of the cloning procedure and linking them with the steps involved in bioinformatics analysis, as described by Hurd (2008). The instructor also has the option of integrating a final written report or oral presentation into the course. For example, students could report on the success of the different wet-lab steps, analysis of their GAPDH sequence, and inferred evolutionary relationships.
Students in our molecular biology course have been taking a hands-on approach to cloning the GAPDH gene from various plant species since 2005. In 2005 and 2006, 100% of the students succeeded in amplifying plant GAPDH; however, in 2007 we saw only 50% success by using tree fern DNA. More than 95% of the groups successfully obtained clones between 2005 and 2007, although additional screening for positive inserts was often required. As a result, 12 GAPDH plastid sequences from plants such as jasmine (accession GQ372996– GQ372998), shamrock (accession GQ372991– GQ372993), coffee (accession GQ372994– GQ372995), wood-sorrel (accession DQ075672), and lime (accessions EF599118, EF601087, and EF613285) have already been published in GenBank, all with students as coauthors.
The Bio-Rad Cloning and Sequencing Explorer Series has streamlined the cloning process and the instructor's prep time, facilitated DNA sequencing, as well as formalized the bioinformatics analysis. The kit has been used in our course for the past 2 yr, with 91% success in amplification of the nested PCR product in 2008 and 92% in 2009. Greater than 91% of the groups successfully obtained clones these 2 yr. This effort has resulted in the publication of four GenBank accessions from dwarf umbrella tree (accession FJ648426), aluminum plant (accession GQ332381), croton (accession GQ332382), and eyelash begonia (accession GQ332383). Plant GAPDH sequences that were isolated from other species in the 2009 semester should be released by the time this article is published. We have observed that the initial and nested PCR primers are fairly robust because amplification was successful with a wide range of angiosperm plants, both monocots and dicots. In some cases, the consensus primers did not seem to amplify nonangiosperms, such as fern and pine tree.
The GAPDH cloning kit has been on the market since fall 2008. Using this kit, at least six other institutions have successfully isolated and published 14 different GAPDH genes from a wide assortment of plant species (Table 4). The adaptability of this kit is reflected in its apparent integration into the curriculum of high schools, community colleges, and universities. With more unique plant GAPDH sequences being isolated and published in GenBank, there will be even greater opportunities to integrate additional phylogenetic approaches in the future.
| Institution | Plant species | Accession no. |
|---|---|---|
| Biotechnology Career Academy (Ellicott City, MD) | Aluminum plant | GQ132134 |
| Biotechnology Career Academy | Petunia | GQ122207 |
| Chapman University (Orange, CA) | Flax | GQ148916 |
| Northwest Nazarene University (Nampa, ID) | Dumb cane | GQ253512 |
| Northwest Nazarene University | Hardy ice plant | GQ219791 |
| Northwest Nazarene University | Venus fly trap | GQ246217 |
| Saint Paul's School (Concord, NH) | Snapdragon | FJ374124 |
| Scott Community College (Bettendorf, IA) | Mint hybrid | GQ241339 |
| Scott Community College | Common balm | GQ241338 |
| School for Science and Math-Vanderbilt University (Nashville, TN) | False pennyroyal | FJ650498 |
| School for Science and Math-Vanderbilt University | Monkshood | FJ649623 |
The Cloning and Sequencing Explorer Series is a sophisticated, multipart, and interdisciplinary exercise that elegantly demonstrates real-world approaches to research. Students gain technical expertise in molecular biology, develop analytical skills, and become more aware of the significance of bioinformatics in the postgenomic era. We feel that this exercise is a unique opportunity for students to be involved in a research process that eventually leads to a tangible end product, which is rather unusual for an educational exercise aimed at the undergraduate, or even high school, level.
ACKNOWLEDGMENTS
Special thanks to Drs. Bryony Ruegg, Sandra Porter, and Melissa Woodrow for collaboration on the GAPDH project. We thank Dr. Laurie Usinger for thorough reading of the manuscript and helpful suggestions. As a student, Carolyn Payne spent many hours on background research. We appreciate all of our former Molecular Biology students at Bellarmine University who have made this exercise worthwhile over the past 5 yr. The collaboration of the Department of Energy-Joint Genome Institute in Walnut Creek, CA, is much appreciated. This undergraduate research-based laboratory project was inspired by National Institutes of Health grant P20 RR16481 from the BRIN Program of the National Center for Research Resources. This manuscript is dedicated to the memory of Ron Mardigian for his commitment to biotechnology education.
FOOTNOTES
Conflict of interest: J.M.L. and D.L.R. have both served with Bio-Rad Laboratories (Hercules, CA) as research consultants on this project and are royalty recipients.



