Enhancing Interdisciplinary Mathematics and Biology Education: A Microarray Data Analysis Course Bridging These Disciplines
Abstract
BIO2010 put forth the goal of improving the mathematical educational background of biology students. The analysis and interpretation of microarray high-dimensional data can be very challenging and is best done by a statistician and a biologist working and teaching in a collaborative manner. We set up such a collaboration and designed a course on microarray data analysis. We started using Genome Consortium for Active Teaching (GCAT) materials and Microarray Genome and Clustering Tool software and added R statistical software along with Bioconductor packages. In response to student feedback, one microarray data set was fully analyzed in class, starting from preprocessing to gene discovery to pathway analysis using the latter software. A class project was to conduct a similar analysis where students analyzed their own data or data from a published journal paper. This exercise showed the impact that filtering, preprocessing, and different normalization methods had on gene inclusion in the final data set. We conclude that this course achieved its goals to equip students with skills to analyze data from a microarray experiment. We offer our insight about collaborative teaching as well as how other faculty might design and implement a similar interdisciplinary course.
INTRODUCTION
Interdisciplinary Mathematics and Biology Education
One of the goals of BIO 2010: Transforming Undergraduate Research for Future Research Biologists (National Research Council, 2003) was to increase the teaching of courses that bridged the disciplines of mathematics and biology. Many papers have been written about the need for increasing interdisciplinary teaching of mathematics and biology and the results and challenges of attempting courses that integrated these disciplines (Steitz, 2003; Bialek and Botstein, 2004; Brent, 2004; Gross et al., 2004; May, 2004; Campbell et al., 2007; Perez-Iratxeta et al., 2007; Knight et al., 2008; Pevzner and Shamir, 2009; Pursell, 2009). Bialek and Botstein (2004) quote Galileo who wrote that “the book of nature is written in the language of mathematics” and suggest that biologists must become “conversant not only with the language of biology but also with the languages of mathematics, computation, and the physical sciences.” We set as a goal to integrate mathematical and statistical concepts into a microarray data analysis course. Analysis of high-dimensional microarray data can be challenging to biologists, especially if they do not have a strong statistical background. In contrast, statisticians often find themselves analyzing biological systems with which they are unfamiliar and with which they have not been trained to interpret. A more ideal system is for the statistician and biologist to jointly analyze the data and transfer their expertise to students. A wet lab microarray course had been taught previously at Rochester Institute of Technology (RIT), and students were undertaking undergraduate research projects using microarrays. Individual independent research projects in which students analyzed a microarray data set using R and Bioconductor had been done. As more students generated microarray data sets and students learned in their course work about the power of microarray analysis, a need arose for a microarray data analysis course. This report discusses how we organized such a course and what we learned from teaching it. We feel that one of the reasons the course was successful was that we had previously established a successful collaboration. We met when Dr. Evans was teaching courses where students generated microarray data and Dr. Tra was teaching courses in statistical data analysis and found that we had a common interest. Initially, students consulted with each of us individually, but we soon realized that more progress would be made if we jointly advised students. The idea to teach a microarray data analysis course where interested students could cooperatively learn how to analyze and interpret microarray data was suggested, and the course described here was the result. Because many biology students are resistant to taking anything beyond the required courses in mathematics and statistics, we focused our course on goals that students had articulated to us.
Many students at RIT are doing undergraduate research projects with microarrays, and these students want to analyze and interpret their data sets. Other students learned about the “microarray revolution” and wanted to be able to use this technology in their future careers. The specific focus of the Microarray Data Analysis course was to help prepare students for careers and graduate study in the biological and mathematical fields as well as to teach students how to analyze data from their undergraduate research projects and other data sets, “mine” the data, and design future experiments. Student-based inquiry was used. Allowing students to choose their projects, analyze, and draw conclusions from their chosen data sets led to student ownership of the course and facilitated active learning. This course also was aimed at fostering and attaining some of the goals articulated in BIO2010 such as an increased emphasis on integrating mathematics and statistics in the biology curricula.
Microarray Technology
The sequencing of whole genomes has changed the research direction in biological sciences and led to the microarray revolution (Butte, 2002; Grünenfelder and Winzele, 2002; Simon, 2003; Brewster et al., 2004; Carpenter and Sabatini, 2004). Microarray experiments ask big questions and generate a large volume of data. DNA microarrays can be used to measure changes in gene expression levels in development and disease, to detect single-nucleotide polymorphisms useful in diagnosing disease predisposition, and to characterize new species of organisms. Genomics, the study of all the genes of a cell, encompasses study of the DNA (genotype), mRNA (transcriptome), or proteins (proteome). In one gene expression profiling experiment, the expression levels of thousands of genes can be simultaneously monitored to study the effects of certain treatments, diseases, or developmental stages on gene expression. For example, microarray-based gene expression profiling can be used to identify genes whose expression is changed in response to pathogens or other organisms by comparing gene expression in infected to that in uninfected cells or tissue. Cancerous tissue can be compared with normal tissue. Over the past few years, microarrays have become the most common tool to obtain repeated measurements of RNA transcripts of genes. Expression-profiling microarrays are artificially constructed grids of DNA in which each element of the grid holds a DNA sequence that is the reverse complement to the target RNA sequence. There are different types of platforms, including two-color microarrays and Affymetrix microarrays. These platforms have become an established technology in molecular biology and are used in an increasing number of laboratories. Microarray use in pharmaceutical research has expanded with applications in basic research for drug target discovery, biomarker determination and validation, and toxicogenomics as well as development of prognostic tests and disease-subclass determinations. Due to the decreasing cost of making microarrays and increasing support for their use in undergraduate education, microarray technology is now accessible for academic use and for undergraduate research. A growing wealth of analysis tools also is available.
Importance of Microarray Data Analysis
Biology is now data-intensive, large, and multivariate. The regular statistics course required by a biology department does not cover the statistical methods appropriate for such data. Many researchers (including biologists) are challenged in analyzing the high-dimensional data produced by microarray experiments. Our task as educators is to equip the students with a strong statistical background and an ability to use adequate statistical methods for the interpretation of the microarray results. Although some courses have been taught successfully to biologists and/or statisticians (Honts, 2003; Heyer et al., 2005; Hardin et al., 2006; Wise et al., 2006), more interdisciplinary courses need to be offered. Microarray techniques are extremely noise-prone, subject to bias in the biological measurement, or both, and a major research area in computational biology involves developing statistical tools to separate signal from noise. The analysis of DNA microarray data requires preprocessing of the data, including quality-control analysis and normalization. Normalization is the first transformation applied to the data expression to adjust the individual hybridization intensities. The goal is to balance the intensities appropriately so that meaningful comparisons can be made. The reasons for normalizing the data includes unequal quantities of starting RNA, differences in labeling or detection efficiencies between the fluorescent dyes used, and systematic biases in the measured expression levels. Unwanted imbalance may also come from dye degradation (Fare et al., 2003) due to high ozone concentrations. Often the microarray experiments were performed with a small number of biological replicates, and this presented problems for detecting differentially expressed genes while avoiding concomitant high false positive rates (Wei et al., 2004). All of these factors presented an opportunity to teach a course in which students learned and used statistical methods to overcome the problems inherent in microarray experiments and then extracted the important information that can be obtained using microarray technology.
Microarray Data Analysis Course: Building on GCAT
The idea of using DNA microarray experiments to teach genomics, computational biology, and bioinformatics concepts for undergraduate researchers started in 1998 when Pat Brown spoke at the American Society for Cell Biology meeting and introduced the audience to the power of microarrays. GCAT (www.bio.davidson.edu/projects/GCAT/gcat.html) founding members were Malcolm Campbell and Mary Lee Ledbetter (Campbell, 2002; Campbell et al., 2006). Educators rapidly endorsed the concept of using microarrays for gene expression studies and the need to introduce students to this powerful new technology (Campbell, 2002; Brewster et al., 2004; Campbell et al., 2006, 2007; Kushner, 2007). Laurie Heyer and colleagues introduced the use of MAGIC Tool (Heyer et al., 2005) written by and with an undergraduate focus so that students could rapidly analyze their microarray data and perform complex analysis. Participants in the GCAT community have access to affordable microarrays, microarray scanners, and free software (MicroArray Genome Imaging & Clustering Tool; MAGIC) for data analysis (Heyer et al., 2005; Campbell et al., 2007). The establishment of GCAT meant that many undergraduate courses were offered, including Dr. Evans' course at RIT. Many of these courses generated interesting data sets that needed to be analyzed. Data analysis can be a major bottleneck for many researchers who may be applying inadequate statistical methods to interpret their microarray results. At the same time, increasingly more sophisticated microarray platforms are introduced along with new analysis methods.
The course was intended to familiarize the students with these advanced methodologies and provide hands-on training on the latest analytical approaches. Students had the opportunity to use real biological data and statistics in biology and were introduced to advanced statistical methodologies and software tools for analyzing and managing microarray data. We think that it is an important part of the training of every student in the biological sciences. This is an area where statistics can be relevant and accessible (Moore, 1997; American Statistical Association, 2005; Hardin et al., 2006). We taught students how to extract meaningful information from a data set with enough confidence to guide future research projects. A solid understanding of the type of inferential statistical method used was also stressed. The course was offered as an elective to biological sciences students in winter quarter 2008. To engage the students in the learning process, an active-learning pedagogy was selected. It has been demonstrated that retention of course material increases in active-learning settings because the students are able to apply their knowledge firsthand (Knight et al., 2008). Students were given a short lecture on a statistical analysis topic and a demonstration of how to use microarray data analysis software. The students then used the software/resources to analyze microarray data. According to Moore (1997), “the most effective learning takes place when content (what we want students to learn), pedagogy (what we do to help them learn) and technology reinforce each other in a balanced manner.” The use of technology such as statistics software is essential to emphasize statistical literacy rather than tedious calculations (Garfield et al., 2002). Students' analytical skills improve by doing practice problems (Hake, 1998). The activities were designed to develop creative thinking, collaborative problem-solving skills, innovation, and the ability to use technology resources to accomplish assigned projects and goals.
Active learning, collaborative work, and use of interactive computer modules were part of the recommendations from expert panels to improve science education (Handelsman et al., 2004; DiCarlo, 2006). These recommendations were incorporated into our course. The purpose and goals of the course were to 1) produce and understand a microarray experiment by doing a “wet lab” (see link in day 2 in online dynamic calendar at http://people.rit.edu/∼yvtsma/index.html); 2) inquire and conduct image analysis with MAGIC or ScanAlyze software (http://rana.lbl.gov/EisenSoftware.htm); 3) organize and import data into R, a statistical software for computing and graphics, for preprocessing, background correction, and normalization; 4) construct data visualization plots; 5) assess statistically significant genes that were “outliers” and thus were over- or underexpressed; 6) incorporate higher-level analysis (Pathway Analysis); 7) read journal papers in which microarray technology and analysis are discussed; and 8) do a project involving analysis of microarray data.
MATERIALS AND METHODS
Course Implementation
Initially, the targeted audiences were third- and fourth-year students from the following disciplines: biology, bioinformatics, biotechnology, biochemistry, biomedical sciences, mathematics, and statistics. The course was opened to any student in related disciplines interested in the subject. It was offered as an elective (four credit hours) and listed as special topics under interdisciplinary sciences. The maximum number of students allowed to register was 10. It was advertised with a flyer weeks ahead of registration to attract many students. A presentation of the course was also done for the math club. The goal was to draw undergraduate students from different disciplines to be trained in the analysis of the data from microarray technology and to be knowledgeable about high-dimensional biology data analysis. Two prerequisites were required to take the class: General Biology or Intro to Biology, and Data Analysis I or its equivalent.
Course Description
No textbook was required. To prepare for the class, reading assignments covering the topic of the day were posted online in a dynamic calendar available at http://people.rit.edu/∼yvtsma/indexm.html where downloadable labs and practicals are also provided as resources. The course was a 10-wk quarter, taught in a team spirit interdisciplinary approach by two instructors, one instructor with expertise in biology and the other instructor with experience in statistics. These courses are encouraged by the respective departments involved. Whenever biological content and understanding were required, the biologist led the teaching and discussion. The statistical materials were taught by the statistician. Both instructors received the same amount of credit for teaching the course. Following are examples of the team-teaching approach; both instructors were present at all class sessions. During the first 2 wk, the biologist instructor introduced gene expression studies, microarray technology and platforms, and the biological aspects of microarrays, and conducted a wet lab microarray experiment with the students. Data were collected. The results of the experiment were used to illustrate the concept of technical versus biological replicates and the need to transform the data into a ratio for finding differentially expressed genes. Discussion on this topic and the data analysis were led by the statistician instructor. The topic on assessing differential expression of cDNA data was illustrated by the Apolipoproteins A-I experiment. The data were from a study of lipid metabolism by Callow et al. (2000). The biologist instructor first explained the use of gene knockout experiments for analyzing specific gene functions. Next, the statistician instructor taught the students how to write a design matrix, normalize the data, and fit a linear model. There were three scientific paper review take-home readings and summary writing (Table 1). Two of the papers addressed the statistical aspects of microarray data. For each paper, a group of students was assigned to do a summary in-class presentation of the paper's major take-home message, followed by a discussion led by either instructor, depending on the paper content. The third and fourth week were devoted to learning the software MAGIC developed at Davidson College by Laurie Heyer and her undergraduate students. The students were able to practice and apply (as part of a daily assignment) the various steps of analysis of microarray data (Figure 1).
| Lecture | It gave insight into how biological knowledge can be generated from microarray experiments and illustrated different ways of analyzing such data. |
| Practical session | Each session (not for grading) demonstrated software and/or resources to analyze microarray data. The practical sessions consisted of computer exercises that enabled the students to apply statistical methods to the analysis of microarray data. Leading questions to evaluate plots were often asked. Critical thinking and interpretation of the results were part of the in-class discussion. Script programs in R were included in these practice exercises. They served as a template to use for computer lab assignments. |
| Computer lab | The focus was on the practical side of gene expression data analysis. After each lecture and practice session, each student worked on a computer lab assignment based on the topic covered. If not done, he or she was allowed to continue outside class time and to turn in the assignment the following class. A daily computer lab included a short report, program scripts, answers to the questions and corresponding required plots. |
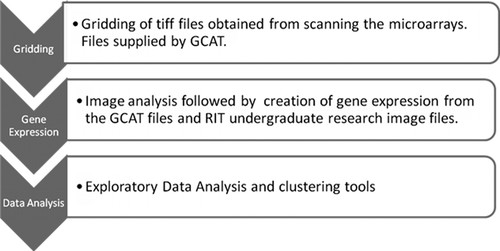
Figure 1. Steps of analysis of microarray data.
Due to MAGIC's current limitations for preprocessing data as well as analysis and comparisons of a significant number of replicates, the last 7 wk were dedicated to learning the software R, a statistical software for computing and graphics. The intent was to acquaint the students with this widely used software and to present some of the important low-level analysis such as normalization and quality control involving preprocessing and flagging data as well as advanced methodology (pathway analysis). Each 2-h class session was a mix of lecture and hands-on activities (Table 2; links provided in the dynamic calendar). Bioconductor packages (Gentleman et al., 2004), along with R, were used for the different stages of analysis for spotted and oligonucleotide microarrays (specific levels of analysis described in Figure 2).
| Lecture topic | Introduction to gene expression studies, microarray technology, and platforms |
| Introduction to R and bioconductor | |
| Image analysis; generating expression data with MAGIC using RIT yeast prion data set | |
| Exploratory data analysis and clustering with MAGIC | |
| Preprocessing cDNA data and Affymetrix arrays with R | |
| Normalization | |
| Differential expression—linear modeling using Limma for both (Affymetrix, two-color microarray) platforms | |
| Gene set enrichment analysis | |
| Classification using R | |
| Activities | Perform a microarray experiment |
| Analyze the microarray experiment | |
| Transforming ratio, finding differentially expressed genes | |
| Articles, reading | Tilstone, C. (2003). DNA microarrays. Vital Statistics |
| DeRisi et al. (1997). Exploring the metabolic and genetic control of gene expression on a global scale | |
| Butte, A. (2002). The use and analysis of microarray data | |
| Group projects | Changes in gene expression during sleep and prolonged wakefulness in the brain of Drosophila |
| Effects of spinal cord injuries on gene expression: gene discovery and pathway analysis | |
| Effect of prefiltering on changes in the gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus | |
| Two-color microarray analysis (dye-swapped) of the epigenetic effects of the [PSI+] and [psi−] phenotype in Saccharomyces cerevisiae | |
| Microarray analysis of Psi+ induced phenotypic changes in yeast | |
| Differential gene expression in anatomical compartments of the human eye using linear models and empirical Bayes method |
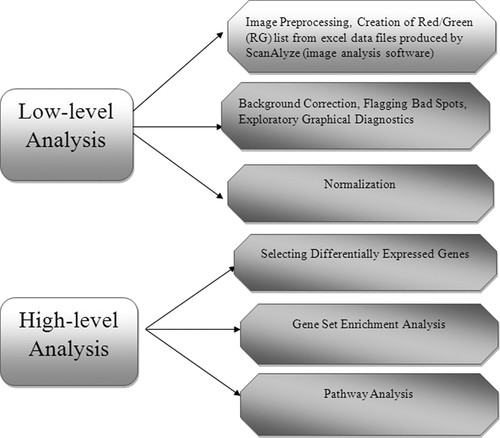
Figure 2. Stages of analysis for spotted and oligonucleotide microarrays.
The gene set enrichment and the pathway analysis were only taught for Affymetrix microarrays by using the available Kyoto Encyclopedia for Genes and Genomes repository (Kanehisa and Goto, 2000).
Student Assessment
Student performance was assessed in three different ways: 1) computer-based labs that were turned in for grading; 2) homework, including summaries of journal papers; and 3) a group project. For the first two assignments, the students were allowed to work in a team of two or three. However, each student was responsible for his or her own writing. Grading was weighted as follows: daily assignments (35%), homework (35%), and project and presentation (30%) (specific in-class routines are given in Table 2).
For the daily computer lab, the focus was on the practical side of gene expression data analysis. After each lecture and practice session, each student worked on a computer lab assignment based on the topic covered that day. If not completed, the student was allowed to continue outside class time and turn in the assignment at the following class. A daily computer lab included a short report, program scripts, answers to the questions, and corresponding required plots. Three homework assignments were required for the quarter. Each student worked on an end-of-term group project of his or her choice. Each group had an in-class presentation of the project the last day of class. Project topics were taken from Microarray Gene Expression Data (www.mged.org) and accepted public microarray data repositories such as ArrayExpress (Brazma et al., 2003). Two of seven students in the class were student researchers who used the data set they produced from microarrays made in their lab. Guidelines on report content and structure were posted online as well as a grading rubric (Table 3) for the PowerPoint presentation.
| Component | Criteria | Exemplary (5 or 4) | Proficient (3 or 2) | Not yet proficient (1) |
|---|---|---|---|---|
| Project proposal | Purpose | Identify topic of interest (without instructor's help). | Identify topic of interest (with instructor's assistance). | Incomplete purpose and too easy to attain topic. |
| Data analysis | Exploratory | Graphs and descriptive statistics with interpretation. | Graphs and descriptive statistics. | Missing or inaccurate graphs or/and descriptive statistics. |
| Use of methods | Demonstrate knowledge of the method by applying it to answer the research question, integrate major concept into the response, show in-depth thinking about the method. | Demonstrate knowledge of the method by applying it to answer the research question, limited thinking about the method. | Do not demonstrate knowledge of the method, no evidence of depth of thinking about the method. | |
| Report | Introduction | Define clearly the study, the objectives, and the research question. | Define clearly the study and the research question. | Missing introduction, no objectives and no research question posed. |
| Data description | Describe the data set (define variables and controls). Explain the data collection process. | Describe the data set and the data collection without details. | Forget to describe the data set and/or the data collection. | |
| Methods of analysis | Describe clearly the selected methods to analyze the data. State the questions of interest. | Describe roughly the selected methods to analyze the data. | Forget to describe the selected methods to analyze the data. | |
| Results | Provide descriptive statistics and graphs. Show relevant R output for the statistical tests. Explain findings clearly—what do the graphs show? | Provide descriptive statistics and graphs. Show relevant R output for the statistical tests. | No descriptive statistics and graphs or graphs are the wrong type. Forget relevant R output for the statistical tests. | |
| Conclusion and discussion | Write conclusion and interpretation in layman's terms. Explain and discuss the significance of findings in the context of the topic. | Write conclusion and interpretation. Explain and discuss the significance of findings in the context of the topic. | ||
| References | List any books, articles, and web pages used, in proper order. | List any books, articles, and web pages used. | Forget to list any books, articles, and web pages used. | |
| Appendices | Data set (or link to the data set). Any computer output. Tables and figures are numbered and captioned. | Data set (or link to the data set). Any computer output, tables and figures. | Forget to give data set (or link to the data set) or/and any computer output, tables and figures. | |
| Presentation | Quality of talk | Clear, eye contact, brief and concise. Enthusiasm and confidence are evident. Presentation fit into 10-min allotment. | Mostly audible and/or fluent on the topic, eye contact broken with audience. Presentation >10-min allotment. | Inaudible and hesitant. Rely heavily on notes, no audience eye contact. Presentation >10-min allotment. |
| Quality of slides | Excellent structure, color, font, animation, original, creative, and holds audience attention. | Well structured, font and resolution appropriate. Somewhat holds audience attention. | Not organized, no color, small font, lack of creativity, and doesn't hold audience attention. | |
| Content of slides | Solutions clearly stated, logical flow of ideas easy to follow, correct spelling and grammar, important graphs included. | Solutions clearly stated, transition and/or flow of ideas somewhat difficult to follow, slides error free, graphs included. | Solution not clearly stated. Unclear conclusion. Transitions and flow not logical. Slides with errors and a lack of logical progression, no graphs included. |
Course Assessment
A midterm anonymous online student clipboard survey (an online survey creation tool for faculty and staff at RIT) was conducted to collect student feedback. There were 19 questions, 12 of which were on a 1- to 5-point Likert scale. The students were asked to reflect on course objectives, content, course design, and assignments and were asked for suggestions for additional content to enhance their learning. An online end-of-quarter evaluation assessed the instructors and the course.
RESULTS
Student Enrollment and Outcomes
The class was composed of four biotechnology majors, two biology majors, and one biotechnology/bioinformatics option major. They were all in their third or fourth year of study. One student from another discipline dropped the class. He was afraid of not doing well based on poor knowledge of biology. This course does indeed require a background in biology and statistics as stated in the course prerequisite. We felt this student could have continued in the course and done well, but the student opted to withdraw.
Students' Assessment Results
The computer labs were designed in a guided manner through which each step of programming and statistical analysis, including the rationale, was explained progressively. Questions were embedded in the labs. A few advanced statistical analysis assignments were presented such as permutation testing for finding enriched pathways. The homework was built with the same structure, summarizing several topics addressed in the labs. The class average for the daily computer labs was 85%, whereas for the homework it was 80%. The difference showed the effect of interaction between the students and the instructors. In-class help was available and reinforced the lecture. It also reflected the level of difficulty of the assignment. The labs contained drill and practice questions assessing knowledge, comprehension, and application, whereas the homework had additional critical-thinking questions involving synthesis, analysis, and evaluation (Bloom, 1956). Students' projects were graded based on a rubric (Table 3) and grades were posted in mycourses, the RIT online course management system. Each component was assigned a maximum of 5 points for a total score of 60 points. Projects were done in groups of two or three, to facilitate collaborative learning, although several students opted to work individually. Each group or individual wrote a paper and also gave an oral presentation. Results showed that three of the projects were judged proficient and four were exemplary. These data indicate that the majority of students were competent and demonstrated evidence of advanced learning as they provided meaningful interpretation of their results. Projects covered different topics and analyzed new or published data sets from two platforms, Affymetrix or two color arrays (project topics displayed in Table 2). In one interesting project, a group of students reanalyzed the change in the gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus. The group showed that filtering data before preprocessing can cause massive data loss. After background correction, normalization, and fitting a linear model, down-regulated genes were recovered. Another project result was the discovery of the top five pathways activated in cells after spinal cord injuries. None of these results were found in the original published papers.
Course Assessment Results
Likert scale questions were used for 12 questions of the midterm survey. Average ratings and related SD for each question were computed (Table 4). Students agreed that course objectives were clear (average rating 4.14) and relevant to their future job interest (average rating 4). The quality of content and activity were rated high, with an average rating of 4.29 (engaging). We had high expectations at the beginning of the course. The below-average (2.29) students' rating of the course design and flexibility suggested a need for adjustment to the students' level of comfort and speed. We recognized that the course was structured in 10 wk with different activities in such a way that there was no room for adjustment. We then tried to accommodate the students and gave extensions when the daily lab work was not completed. Regarding activities being helpful, the average rating was 3.28, whereas being ample enough to reinforce understanding was 4.28. The average number of hours to complete an assignment (daily computer lab not done in class) and homework was 2 h.
| Questiona | Avg. rating | Avg. rating SD |
|---|---|---|
| The objectives of the course were stated clearly | 4.14 | 0.38 |
| The objectives of the course are relevant to my future job interests | 4.00 | 0.58 |
| The course content and activities are engaging | 4.29 | 0.49 |
| The design is flexible enough for me to move around at my own pace | 2.29 | 0.95 |
| There are ample number of activities | 4.29 | 0.76 |
| The placement of activities makes sense | 3.57 | 0.79 |
| The activities helped to reinforce my understanding of the content | 3.28 | 1.11 |
| The course content is covered to an appropriate degree of breadth | 3.28 | 0.76 |
| The content is clearly explained | 3.43 | 0.79 |
| The assignment directions are clear | 3.38 | 0.74 |
Although six of the students had previously taken a computer programming course, learning and writing programs in R was a challenge. One of the student comments was “The course is hard because it involves programming and statistics. It was harder than I thought, but I'm having fun learning about it. I wouldn't recommend taking more than one higher-level course if people decide to enroll in this class because there's just too much work needing to be done outside class. Perhaps fewer materials should be covered with extra time on the programming aspect.”
Overall, the small average rating SD suggests that there is a consensus on the students' opinions on each question. The course evaluation (Table 5) showed that five of seven students always attended class. The majority spent between 10 and 15 h a week (outside of class time) on this course. When asked “How much did you learn in this course?,” one student responded a moderate amount, three responded a lot, and three responded an exceptional amount. In the BIO2010 report, Richard Feynman quoted “… The best teaching can only be done when there is a direct individual relationship between a student and a good teacher …. It's impossible to learn very much by sitting in a lecture …” To the question “It was evident that the instructor encouraged student involvement” five students agreed and two strongly agreed. Based on overall learning experience, six of seven students would recommend the instructors to others.
| Question | Occasionally | Usually | Always |
|---|---|---|---|
| How often did you attend this class | 0% | 28.6% | 71.4% |
| 0–5 h | 5–10 h | 10–15 h | |
| Hours per week, other than class time, spent on this class | 0% | 42.9% | 57.1% |
| Disagree | Agree | Strongly agree | |
| The instructor made the expectations for the course clear | 0% | 85.7% | 14.3% |
| The instructor presented the material clearly | 0% | 85.7% | 14.3% |
| The instructor set a reasonable pace for the course | 71.4% | 14.3% | 14.3% |
| Attending class helped me learn | 14.3% | 28.6% | 57.1% |
| The instructor answered questions effectively | 0% | 57.1% | 42.9% |
| The instructor encouraged student involvement | 0% | 71.4% | 28.6% |
| Sufficient graded feedback was provided | 0% | 57.1% | 42.9% |
| Assignments helped in understanding the material | 14.3% | 42.9% | 42.9% |
| Based on my overall learning experience, I would recommend the instructor to others | 0% | 85.7% | 14.3% |
| A moderate amount | A lot | An exceptional amount | |
| How much did you learn in this course | 14.3% | 42.9% | 42.9% |
Lessons Learned
We believe that the daily activities (practical session and in-class computer lab assignments) contributed the most to student learning. The students were able to apply the methods appropriate for their project. The other thing that contributed to individualized student learning was the writing of a rough draft of the project. By looking over and critiquing a student's rough draft, we were able to lead the student in the right direction and correct any misunderstandings. As a result, the student got the chance to modify his or her research question. During the 10-wk instruction, the biologist instructor was responsible for 3 wk of materials and participated (for the 7 wk left) in class discussion related to her expertise. One of the advantages of team-teaching is the complementarities of expertise provided to the students for each covered topic. As a team, course objectives, sequence of topics, and materials to be covered were set together. The instructors deepened their friendship and collegiality through their shared responsibility. In the midterm survey, one student requested a full analysis of a data set and when other students agreed that this would strengthen their understanding, we implemented such a complete analysis. The quality of learning was enhanced as a result of these changes. Due to different student backgrounds and abilities, we realized that the majority of the students were not able to finish the computer lab assignment in class; as a result, the due date for the computer lab assignment was changed to the next class period. Student comments from the final week survey were constructive. Representative comments are listed below.
“Assignments are a lot. Every class, we have a lab report due and normally the students can't finish the lab on time. We also have practices and they were done in class, but sometimes students do not really understand the practices, which makes it harder to do the lab afterward.
“Maybe slow it down a little bit. In one week, we have 2 labs and one weekly assignment and since the course is hard, students spend a lot of time outside class to find alternative sources online or wait for office hours etc. But all the assignments were helpful.”
“Instructor was approachable. This quarter, I spent most of my time outside class learning the material and doing the labs and weekly assignments. For biology students, this course is quite hard so I wouldn't recommend students taking this course if they have more than one higher-level course in biology.”
“I asked questions about things I only kind of understood and the responses were very helpful in allowing me to continue on my projects. I think what I didn't understand came from just not being familiar with the language syntax most of the time.”
“Good organization of the course material, but she could have presented the materials better. We have many things to learn and maybe because of that, she went really fast.”
“This course seems hard to teach because the students have very variable backgrounds in statistics and general computer skills. Also, you tried to cover MAGIC, 2-color microarrays, and Affymetrix arrays. Just a lot of material; I am surprised we got through as much as we did.”
The students had an appropriate required biology background, but not the programming skills required to conduct the analysis. Based on these comments and our observations, we propose the following modifications to facilitate learning the second time this course is offered:
Cover fewer materials, e.g., spend more time on the “how to do it” programming part and less on different methods for discovering differentially expressed genes.
Space out lab reports to reduce the speed of new materials delivery and focus on making the materials easy to grasp.
Increase instructor and student interaction by having feedback discussions after each practical.
Present the materials better by rewriting them. Try to use one data set from start to finish, but also offer examples from other data sets for illustration.
Have both instructors support the students in the programming part or have the support of a graduate teaching assistant during lab activities.
Keep open the communication lines between the instructors and evaluate weekly how the course is progressing.
Keep the class size small.
DISCUSSION AND CONCLUSIONS
Reflections on a Cross-Disciplinary Team
There were two challenges we faced while working as a cross-disciplinary team. The first challenge was the time invested in acquiring knowledge of the subject and research culture of the unfamiliar discipline. The culture, way of thinking, and problem-solving approach differed between the disciplines of mathematics and biology. These differences affected (positively and negatively) the collaboration. The language and the background were different. The work of one discipline implied certain conclusions that could be drawn, but the other discipline felt such conclusions might not be acceptable because they neglected to address a critical dimension of the problem. The second challenge was acquiring a collaborative attitude. Good communication, willingness to try to understand another point of view, and respect for each other's expertise were the keys to success. A lot of education took place to the benefit of both parties. The following example illustrates the differences between how a statistician and a biologist evaluate microarray data. When a statistician compares two conditions (treatment vs. control), a gene is differentially expressed if its expression level changes systematically, confirmed by a small p-value, regardless of the magnitude of the difference. A test statistic determines the change in gene expression relative to the underlying noise in the gene. Taking variability into account is important. For a biologist, the magnitude of the difference is important. Typically, a twofold change or more is what is further studied. A fold-change is defined in two ways in the literature: as the ratio of the mean control and mean treatment observations and as the difference of the mean log control and mean log treatment data. This biological consideration looks at an absolute change in gene expression, ignoring noise. The difference between the two languages for the same goal is obvious. Mathematicians were trained to give precision and rigor to the concepts and results. Biologists knew the biological system of interest and wanted to investigate and confirm the microarray results using other methods such as real-time reverse transcriptase-polymerase chain reaction. Further biological investigation was difficult if the observed changes in gene expression were small. The biologist also wanted to incorporate the findings into what was already known and was sometimes uncomfortable with small fold-changes even if they were very significant statistically. Discussions between the statistician and the biologist would ensue about what results should be emphasized. In this back-and forth encounter, progress was possible. In the end, both sides agreed that both p-value and fold-change are important to determine differential gene expression. Both criteria can be used in designing future experimentation. Furthermore, each side was able to develop some tolerance for trying to understand an unfamiliar field and point of view and expressed respect for the other culture. For example, when the arrays did not have good quality based on diagnostic plots, more arrays were produced until a good set was obtained. Trust was essential when the results were not as expected. Efforts were made to understand each other's expertise and point of view to make sense of what was found.
Advice for Instructors Wishing to Teach a Similar Course
A course such as this could be implemented readily in other settings as an upper-division elective course. From our experience, we suggest that the course be elective and not required so that students self-select to take the course. Students need to be comfortable with computers and familiar with using computer programs. Students at RIT in the bioinformatics program had the background to do well in the course as did the biotechnology and biology students. These students were using microarrays in their classes or in their undergraduate research projects. Instructors would have to modify the course if it were taught to a less motivated and/or more general audience. The instructors also would have to decide on what tools to use for the analysis. The R and BioConductor software used in this pilot course are challenging and represent state-of-the-art technology. The different available packages allowed students to look at the data from several points of view and to apply the statistical method in the related package. There are journals that require investigators to make their original data sets available on a website. Links for these websites are provided in our online course calendar. Open source software packages such as R, Bioconductor, and MAGIC are widely available as well as tutorials for using them. With these resources, it should be possible for a course like the course described above to be designed and implemented by other instructors. The benefits of team teaching can be many. The gains outweigh the challenges. The findings stemming from our cross-disciplinary team approach were rewarding academically and personally. We also learned to appreciate the views of the other discipline and enhanced our own understanding of real-world problems. The study of microarrays provides opportunities for the application of mathematical models. The results helped us to think about new perspectives and challenged any incorrect concepts. For the students, the course and related research were a perfect way to introduce them to this team-based approach for their future careers. Communication skills were also gained for all participants.
ACKNOWLEDGMENTS
This work was inspired by GCAT. It was supported by the Provost Learning Innovation Grant at RIT.



