Genome Science: A Video Tour of the Washington University Genome Sequencing Center for High School and Undergraduate Students
Abstract
Sequencing of the human genome has ushered in a new era of biology. The technologies developed to facilitate the sequencing of the human genome are now being applied to the sequencing of other genomes. In 2004, a partnership was formed between Washington University School of Medicine Genome Sequencing Center's Outreach Program and Washington University Department of Biology Science Outreach to create a video tour depicting the processes involved in large-scale sequencing. “Sequencing a Genome: Inside the Washington University Genome Sequencing Center” is a tour of the laboratory that follows the steps in the sequencing pipeline, interspersed with animated explanations of the scientific procedures used at the facility. Accompanying interviews with the staff illustrate different entry levels for a career in genome science. This video project serves as an example of how research and academic institutions can provide teachers and students with access and exposure to innovative technologies at the forefront of biomedical research. Initial feedback on the video from undergraduate students, high school teachers, and high school students provides suggestions for use of this video in a classroom setting to supplement present curricula.
INTRODUCTION
With the prevalence of genome science comes an increasing need for educational materials to support student learning related to this topic. Genome science will affect students' futures by leading to medical advances (Subramanian et al., 2001; Collins and McKusick, 2005), raising new ethical questions for society (Meslin et al., 1997; Sharp et al., 2004), and possibly directly affecting students' careers if they continue in science. This article describes a new electronic educational resource designed to help introduce students to genomics by showing them how genome sequences are obtained. Washington University School of Medicine Genome Sequencing Center (WU GSC) and WU Department of Biology Science Outreach created a four-part video tour depicting the processes involved in large-scale sequencing titled “Sequencing a Genome: Inside the Washington University Genome Sequencing Center.” This 30-minute tour follows the steps in the sequencing pipeline and is interspersed with animated explanations of the scientific procedures used at the facility. Additional supplemental materials include career interviews with the staff, a glossary, and hands-on activities.
Although the WU GSC regularly gives on-site tours, most students cannot visit WU GSC, nor do they have an opportunity to learn the steps required to produce a genome sequence. The video described here provides students with an introduction to the genome-sequencing process. The target audience for this project ranges from advanced high school biology students to college undergraduate students. The two main goals for this project were to: 1) increase students' awareness of what goes on at a genome sequencing center, and 2) make the video tour interesting and understandable for our target audience.
This project is also part of a long-term program to develop materials to help students understand genomics, including sequencing and other high-throughput technologies designed to gather data on a genomic scale. Curricular tools have been, and continue to be, developed to accompany this video that explain major steps in the genome-sequencing process. The video tour is freely available (as a series of short videos) for download, accompanied by supporting materials on our Web site ( http://www.nslc.wustl.edu/elgin/gsc.html).
With open access to genomic information available on the Web, the Internet has become the primary source for information about genome projects and the main place to access new educational materials about genomics (Guttmacher, 2001). These educational materials range from text-based tutorials to more interactive formats. Over 30 research centers around the globe currently contribute to genome-sequencing efforts, and many of these centers' Web sites contain text-based tutorials and teaching tools available for online learning about genome sequencing ( http://www.ornl.gov/sci/techresources/Human_Genome/research/centers.shtml). Some examples of Web-based interactive tutorials in genomics include resources available through DNA Interactive (DNAi; http://www.dnai.org, part of the Dolan DNA Learning Center, http://www.dnalc.org), and the Human Genome Project Education Kit from the National Human Genome Research Institute (NHGRI) ( http://www.genome.gov/Pages/EducationKit/) (NHGRI, 2003). There are also several examples in the literature of curricula developed to introduce students to genomic information on the Web (Palladino, 2002; Campbell, 2003; Centeno et al., 2003; Bednarski et al., 2005). However, none of these resources provides a video tour that explains the human component of producing a genome sequence.
The project described here is unique in that it introduces students to the technology, equipment, and people who perform high-throughput sequencing while describing the science at each step. The tour follows the “production sequencing pipeline,” which is the series of steps taken to produce the raw sequence component of a genome project, beginning with the cloned DNA of an organism and ending with a complete genome sequence. This video tour complements existing Web-based educational materials in genomics and may be used to help introduce students to large-scale sequencing. The sequencing strategy shown is that in current use at the WU GSC for large, complex genomes and includes generating a BAC-based map of the genome as well as shotgun sequencing and computer-based assembly. Simple genomes can now be sequenced by Whole Genome Shotgun alone; that approach can readily be understood as simply scaling up the process used for sequencing a BAC. The Sanger dideoxy sequencing method is used exclusively; as alternative methods come into use (e.g., Margulies et al., 2005), we plan to add segments describing these.
PROJECT DESIGN
This multimedia project contains several parts. First, there is the video tour of the WU GSC that gives viewers insight into the physical space of the sequencing center and introduces them to robots and other equipment involved in high-throughput approaches. Animations appear throughout the video to explain the more detailed and molecular scientific processes. Other sections include career interviews with WU GSC employees, an animated explanation of the chemistry of dideoxy sequencing, and a glossary of genomics terms. All of these can be viewed from the Web site ( http://www.nslc.wustl.edu/elgin/tour/html/gsc.htm) in Quicktime and are available for download at http://www.nslc.wustl.edu/elgin/gsc.html. Other formats available on request are VHS, CD-ROM, and DVD. Please see“ Technical Design” in the Appendix for more details.
Video Tour
For the tour, we recruited two undergraduate students from the Biology and Engineering Departments at Washington University. The students, Libby and Bryson, follow the WUGSC tour guide, Andrea, who answers their questions as they make their way through the WU GSC. The students work their way through the Center following the production pipeline. During the tour, each step in the pipeline is described in detail, showing the equipment and people involved. Detailed animations integrated into the video demonstrate the biological and chemical steps that are performed while Andrea provides a verbal explanation.
The WU GSC is divided into “cores.” Each core has a different role that is essential for the accurate determination of a genome sequence. Figure 1 shows an outline from the video that describes the different “cores” of the center. The tour begins in the “Media and Materials Core,” where reagents and media are prepared, then progresses through the “Production” cores, where the DNA sequence is eventually obtained, and the “DNA Sequence Management” cores, where the sequence is analyzed and annotated. In the final step, the sequence is deposited into a public database. Figures 2 and 3 show examples of how, for each new core the students visit, the video provides a view of the technicians using specialized equipment for that area, a close-up view of the equipment, and animations describing the manipulation and/or molecular process that is occurring at that step.
Chemistry of Sequencing
Because this video was created for audiences with varying backgrounds and different educational goals, the animation segment titled “Chemistry of Sequencing” was made optional and can be viewed separately from the tour. A snapshot of this segment is shown in Figure 4. In this segment, the modified Sanger sequencing method used by the WU GSC is explained and represented at the level of chemical bond formation, with the sugar-phosphate groups of the nucleotides illustrated in a ball-and-stick representation. During the tour, the sequencing reactions are described in more general terms. Including the segment “Chemistry of Sequencing” separately allows instructors to choose the level of explanation of DNA sequencing that is appropriate for their students.
Careers
In the career segment, viewers can gain insight into the work of the people at the WU GSC, how these employees view their roles, and how their education and experience led them to their positions. A cross-section of people was chosen to illustrate the diverse positions within a sequencing center and the racial and gender diversity of genome scientists and staff. The interviews can be viewed with the tour or shown as a stand-alone segment.
Figure 1. This figure is used in the video to illustrate the sequence of the production pipeline and the division of the pipeline into the WU GSC“ cores.” In the tour, the students begin in the “Media and Materials Core” and end in the “Finishing” and“ Analysis” cores, as shown here. This figure is also provided as a handout for students to have while viewing the video and is available in the Appendix.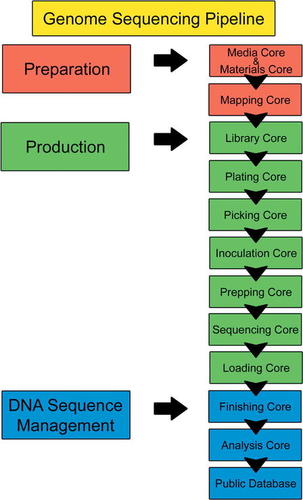
EDUCATIONAL CONTEXT
The video was created for advanced high school and undergraduate biology students and assumes students already have a basic understanding of gel electrophoresis and the concept of recombinant DNA. The processes of bacterial transformation, polymerase chain reaction (PCR), and DNA sequencing are described in the video with animations, but previous or accompanying laboratory experience with these processes will be helpful. There are several online resources for labs with this focus. Some suggestions are provided in the Appendix.
Figure 2. Pictures from the video that show a WU GSC scientist working with a robot, a close-up of the robot, then an animation that reviews the procedure. (a) A WU GSC technician, Bryson, Libby, and Andrea talk about a robot in the“ Picking Core.” (b) A close-up view of the robot picking a colony of bacteria. (c) One frame of an animation that reviews the steps from the transformation to inoculation of a liquid culture of the transformed bacteria. This frame shows the step performed by the robot seen in “b.”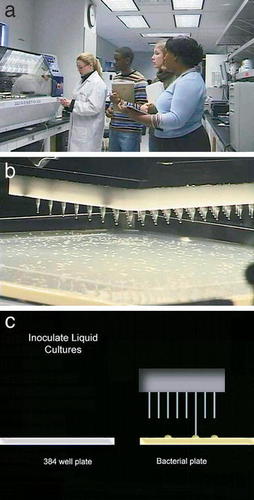
In addition, we have created hands-on paper activities that can be used to emphasize important concepts in the video. These activities are designed for the high school classroom and require no special equipment. Students use paper cutouts of DNA sequences, nucleotide bases, and restriction fragments to simulate the scientific concepts. The “Golden Path” activity allows students to use data from a restriction enzyme digest gel to overlap cloned DNA fragments that contain the same restriction sites, then choose the most efficient “path” of DNA fragments for sequencing. In“ Paper PCR,” students take the role of Taq polymerase and use paper nucleotides to understand how a fragment of DNA can be amplified through several rounds of temperature cycling. “Paper Terminators” is designed to follow “Paper PCR” and illustrates how dye terminators are used in the sequencing reaction. Additional activities, including one illustrating assembly of the final genomic sequence from fragment sequences, will be added. These activities are freely available for download from the video Web site ( http://www.nslc.wustl.edu/elgin/gsc.html).
The video assumes that the instructor has already generated student interest in DNA sequencing through discussion or case studies on the uses of genomic information. One illustration is included: in a short video segment a student talks with a microbiologist to see how sequencing of uropathogenic E. coli suggests how the bacteria cause disease and points toward better therapy.
Figure 3. Pictures from the video that show (a) a WU GSC scientist working with one of the sequencing machines. (b) A close-up of the capillary electrophoresis system used by the sequencing machine. (c) One frame of an animation that shows how the information obtained from capillary electrophoresis is detected and transmitted to a computer that uses the information to create an electropherogram.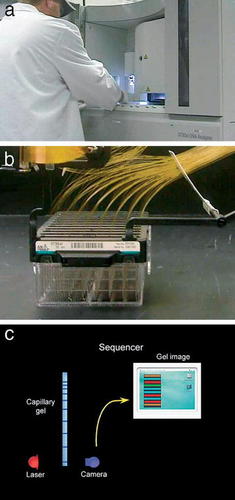
FEEDBACK
We obtained feedback on the video tour from undergraduate students, high school teachers, and high school students. Each group was asked to respond to statements about the video by circling 1-5, with “1” representing no agreement with the statement and “5” representing complete agreement, an approach very similar to the Likert scale for measuring attitudes (Likert, 1932). Each group was shown the video of the tour, but the supplemental sections on“ Chemistry” and “Careers” were not shown to these test groups. The feedback forms were similar for each group and are available in the Appendix and on the Web site ( http://www.nslc.wustl.edu/elgin/gsc.html).
Figure 4. Two frames from the “Chemistry of Sequencing” animation. (a) An overview of the dye terminator reaction in DNA sequencing. Polymerase is shown as an orange oval traveling along the growing DNA strand. The bases are color-coded rectangles. The dye terminators are shown as bases with a brightly colored circle at one end. (b) A growing DNA strand in a sequencing reaction. The arrow points to a 3′ hydroxyl group of a ribose (oxygen is shown as a yellow sphere, and hydrogen is shown as a pink sphere). A nucleotide is shown approaching the 3′ hydroxyl. In the next step (not shown here), the new bond that is formed between the 3′ hydroxyl group and the 5′ α-phosphate of the nucleotide is shown.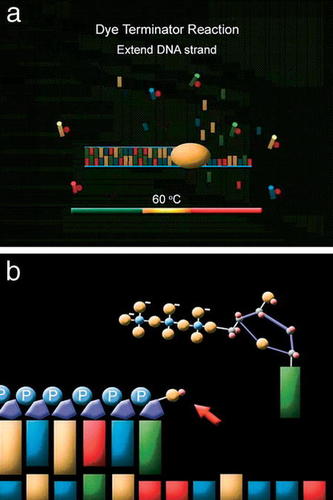
College Undergraduates
Table 1 shows feedback data from the undergraduate students. These students were enrolled in a biology introductory course sequence and were majoring in biology or a closely related field of study. These students were either freshman- or sophomore-level undergraduates who had completed a semester of molecular biology lecture and laboratory that included an introduction to DNA structure, bacterial genetics and cloning procedures, and DNA sequencing. Some of the material in the video was new, and some was a review for these students. Their responses showed that the video was easy to follow (3.9) and helped them understand what went on at a genome-sequencing center (4.4), which were our main goals for the project.
| Question | Average scorea |
|---|---|
| The video was easy to follow. | 3.9 ± 0.8 |
| The video was interesting. | 3.4 ± 0.9 |
| The video helped me understand what goes on at a genome-sequencing center. | 4.4 ± 0.8 |
| The video helped me understand the science of genome sequencing. | 4.0 ± 0.7 |
High School Teachers
Another target audience for this video was advanced high school biology students. Feedback was obtained from high school biology teachers who currently teach genetics in their courses. The teachers were asked if they felt that the video would be useful in their classes and understandable and interesting to their students. The responses indicated extent of agreement to a series of statements on a scale of 1-5 as above. These data are shown in Table 2. Teachers agreed with statements that the video was at an appropriate level for their students and that their students would find the video interesting (averages of 4.0 and 3.8, respectively). On average, teachers also felt the video would help their students understand the science of genome sequencing (4.4).
| Question | Average scorea |
|---|---|
| I think the video is at an appropriate level for high school students. | 4.0 ± 0.9 |
| I think high school students would find the video interesting. | 3.8 ± 1.1 |
| I think the video would help high school students understand the science of genomic sequencing. | 4.4 ± 0.8 |
| Additional questions on use | Average scorea |
| I think high school students would need preparation before viewing the video. | 4.3 ± 1.2 |
| I think high school students would need the Genome Sequencing Pipeline diagram as a handout while viewing the video. | 3.9 ± 1.3 |
| I would need to stop the video periodically during viewing to explain concepts to my high school students. | 4.4 ± 0.8 |
| I would consider showing the video to my high school students. | 4.0 ± 1.1 |
The feedback also showed that teachers agreed they would need to prepare their students before having them view the video and would stop the video periodically to offer further explanation of some of the concepts to their students. Teachers also felt their students would need a handout of the Genome Sequencing Pipeline diagram to refer to as they watched the video (available in Supplemental Materials).
During further discussions, the teachers indicated that the video would be most appropriate for their advanced biology students and that supplemental materials and lab activities would help their students understand some of the material covered in the video. Based on this feedback, we have made several suggestions for classroom and laboratory activities that would help provide the needed context and background for this video in both high school and college classrooms (see “Educational Context”).
High School Students
High school students from a St. Louis school also provided feedback on the video. This group of high school students were currently enrolled in a general biology course but had no or limited exposure to genetics and genomics. As shown in Table 3, these students were not as likely to find the video easy to follow or as interesting as the undergraduate students. Although further assessment is needed to confirm why these students were less likely to find the video interesting, one possibility is that they had less background in genetics and little exposure as to how genomic information is being used to study problems of interest. (The segment “Exploring a Pathogenic Bacterium” has been provided to help address this issue.) These results agree with the comments from high school teachers that the video may be better suited for advanced students or as part of a larger genetics curriculum.
| Question | Average scorea |
|---|---|
| The video was easy to follow. | 2.4 ± 1.4 |
| The video was interesting. | 1.3 ± 1.5 |
| The video helped me understand what goes on at a genome-sequencing center. | 3.0 ± 1.4 |
| The video helped me understand the science of genome sequencing. | 2.5 ± 1.4 |
CONCLUSIONS
Initial feedback from undergraduate students shows that they found the video interesting and understandable. With this video, undergraduate students can watch lab procedures that they may have performed on a small scale be scaled-up to provide the sequence of an entire genome. For high school students, this tour introduces them to genome science and to career opportunities that are available in this area. The feedback from high school teachers shows that they are enthusiastic about showing the video to their advanced classes and that the supplemental activities will be important in helping the students understand the science that is explained in the video. The feedback from high school students also suggests that providing background materials in genetics and genomics to the students before they view the tour will be important. The curriculum materials created to accompany this video along with other resources listed in the Appendix can help provide the needed background.
In recent months, the target audience has expanded to include WU GSC employees and the general public. Because not all of the staff at the WUGSC have science backgrounds, the video has been used as a training tool for new employees and for those who take part in the internal “introduction to biology” courses taught by WUGSC staff. Current employees can use the video as a way of becoming familiar with each core group and of learning how each of these groups contributes to the overall sequencing of a genome.
Additionally, the video is being used as a way to introduce visitors to the WU GSC before they participate in an actual tour. Because there is so much new information that is covered, tour guides at the WU GSC are finding it helpful to show this video to groups before they embark on the tour. The video helps introduce visitors to the concepts of high-throughput sequencing and its applications and to career opportunities in the field of genomic science and provides them with an overview of the WU GSC before taking the tour.
This video program is part of a long-term project of developing materials to help students understand the science of genome sequencing. As the video tour and curriculum tools experience wider use by broader audiences, obtaining additional feedback will be important, and we have included a form on our Web site to collect viewers' comments. This feedback will be used to guide future curriculum development and assessment activities in order to create effective tools that help students develop an understanding of genome sequencing and how this new science may affect their lives.
SUPPLEMENTAL MATERIALS
Web site for accessing the video segments, glossary, video script, and hands-on activities: http://www.nslc.wustl.edu/elgin/gsc.html.
APPENDIX
RELATED CURRICULAR RESOURCES
DNA Structure and Introduction to Bacterial Transformation
Washington University Science Out reach Modern Genetics Curriculum. Teacher and student pages are available for download from the following Website: http://www.so.wustl.edu/science_outreach/curriculum/genetics/download.html.
| Lab names | Chapter and section | Topic |
| Spooling DNA | Ch. 1, Sec. B | Isolating DNA |
| DNA Structure | Ch. 1, Sec. C | Building a DNA model |
| Shine On! | Ch. 1, Sec. E | Bacterial transformation |
DNA Isolation, PCR, and Gel Electrophoresis
Alu Fingerprinting. Dolan DNA Learning Center, Cold Spring Harbor Laboratory. Materials description and laboratory procedure are available at: http://www.geneticorigins.org/geneticorigins/pv92/aluframeset.htm.
Introduction for Alu lab at: http://www.geneticorigins.org/geneticorigins/.
Background on Genome Sequencing
WUGSC: http://genome.wustl.edu/
NHGRI: http://www.genome.gov/
NIH: http://www.ncbi.nlm.nih.gov/About/primer/
DOE: http://www.ornl.gov/sci/techresources/Human_Genome/publicat/primer/toc.html
Online Education Modules
DNAinteractive: http://www.dnai.org/
NHGRI: http://www.genome.gov/Pages/EducationKit/
Technical Design
Approximately seven hours of video footage were shot over a two-day period. Production equipment included a Sony GSR-200 DVCAM camera with lighting equipment and a collection of microphones.
In postproduction, the footage was digitized into a Macintosh-based Media100 nonlinear editing system. This allowed the editor to create sequences with video, graphics, animation, titles, and narration all within a digital realm. A musical score was created by a member of the WU Music Department and integrated into various scenes. The animation sequences were assembled with hundreds of images in Adobe Photoshop and integrated throughout the video. Once completed, the 30-minute video was exported to VHS for distribution on videotape. It was also compressed and rendered as Quicktime and MPEG2 files for integration into a Video CD-ROM and DVD.
The CD-ROM was developed using Macromedia Director. Video and animated sequences were presented individually to allow nonlinear exploration of the material. Users begin by selecting one of three topics on the main menu, including: VIDEO TOUR (the video walkthrough), CHEMISTRY OF SEQUENCING (the animated segment illustrating Sanger sequencing with voice-over narration), and CAREERS (video interviews with GSC staff). Clicking on a selection takes the user to that section of the program. At the bottom of each page is a secondary menu with the following links: GLOSSARY, ABOUT, LINKS, CREDITS, and HELP.
To accommodate those with DVD players, we created a third version of the program using Apple's DVD Studio Pro. The DVD contains all the elements of the CD-ROM, but the large-capacity format allowed us to minimize compression and maintain the picture quality of the original video.
A final version of the program was created for the Internet. The navigation of the CD-ROM was converted to html, and the video segments greatly compressed to decrease their file size. All files were then uploaded to a WU streaming server. This copy is available at http://www.nslc.wustl.edu/elgin/gsc.html.
We created different versions of the program to reach as many end users as possible. The CD-ROM includes a cross-platform design that allows it to run on both Macintosh and PC systems. To view the video on the CD-ROM, users must have Quicktime Player installed. To access the DVD version, users need only a DVD player. To view the Internet version, users must have installed a browser and Quicktime Player. External speakers are recommended for all versions to fully experience the video segments.
Forms and materials for download (pdf files):
FOOTNOTES
Monitoring Editor: A. Malcolm Campbell
§ Co-first authors.
ACKNOWLEDGMENTS
Gabriella Farkas created all the animations for the video, including those represented in Figures 1, 2, 3, 4 of this paper. William Lenihan provided music for the video. Elizabeth Slawson and Bryson Cole were the students featured in the tour. Cherry Chambers, Jasna Despot, and Shannon Lewis provided production support. Juanita Chambers and Elizabeth Slawson helped write the script for the tour. We would also like to acknowledge Jere Confrey, Scott Hultgren, and employees of the WU GSC for participating in the making of the video.
This project was supported in part by Howard Hughes Medical Institute (HHMI) grant 52003904 to Washington University for S.C.R.E., HHMI Professor, and by NHGRI through its support of the WU GSC outreach program.