pClone: Synthetic Biology Tool Makes Promoter Research Accessible to Beginning Biology Students
Abstract
The Vision and Change report recommended genuine research experiences for undergraduate biology students. Authentic research improves science education, increases the number of scientifically literate citizens, and encourages students to pursue research. Synthetic biology is well suited for undergraduate research and is a growing area of science. We developed a laboratory module called pClone that empowers students to use advances in molecular cloning methods to discover new promoters for use by synthetic biologists. Our educational goals are consistent with Vision and Change and emphasize core concepts and competencies. pClone is a family of three plasmids that students use to clone a new transcriptional promoter or mutate a canonical promoter and measure promoter activity in Escherichia coli. We also developed the Registry of Functional Promoters, an open-access database of student promoter research results. Using pre- and posttests, we measured significant learning gains among students using pClone in introductory biology and genetics classes. Student posttest scores were significantly better than scores of students who did not use pClone. pClone is an easy and affordable mechanism for large-enrollment labs to meet the high standards of Vision and Change.
INTRODUCTION
For more than 100 yr, white papers and studies have called for improved biology education at the undergraduate level (McEwing, 2003). More recently, BIO2010 (National Research Council [NRC], 2003) and Vision and Change (American Association for the Advancement of Science, 2011) enumerated the need for original research experiences in undergraduate science courses. The purpose of real research experiences is to improve science education while also increasing the number and diversity of scientifically literate citizens. In addition, the redesigned Advanced Placement (AP) Biology curriculum pushes teachers to provide open-ended and realistic laboratory experiences for their students (College Board, 2011; Wood, 2002; Barsoum et al., 2013). Unfortunately, faculty who teach undergraduates or high school students were not given any additional training, laboratory resources, or time during the day to meet the new demands. Therefore, there is a great need for new laboratory modules that meet the high expectations of educational reform without placing unreasonable demands on faculty time or institutional budgets.
We have developed a laboratory module called pClone that uses typical laboratory equipment, requires minimal preparation time, and meets the increased demands placed on faculty and departments. The development of pClone began in our undergraduate synthetic biology research programs. Synthetic biology integrates the disciplines of biology, mathematics, computer science, chemistry, and engineering (Khalil and Collins, 2010; Biotechnology and Biological Sciences Research Council [BBSRC], 2011; Kahl and Endy, 2013). Using synthetic biology and pClone has the added benefit of bringing students into an area of cutting-edge scholarship as called for in national biology education reforms (McEwing, 2003; NRC, 2003). A shared vision in the calls for education reform is that biology curricula should reflect current areas of research rather than harken back to classic but static fields, such as anatomy and histology (NRC, 2003; AAAS, 2011). Synthetic biology is well suited for undergraduate research (Eckdahl et al., 2010; Campbell et al., 2012), is practiced by a growing community of undergraduate educators (Wolyniak et al., 2010), is attracting federal and corporate funding, and provides promising job opportunities for future scientists. After years of training undergraduates to conduct synthetic biology research (Haynes et al., 2008; Baumgardner et al., 2009; Pearson et al., 2011; Sawyer et al., 2012), we are familiar with the best methods to use with inexperienced students. We have developed pClone as an inexpensive, student-friendly suite of three plasmids that enables novice students to successfully conduct original promoter analysis research. This paper describes our “action research,” which documents learning gains by students using pClone and details the simple procedures by which pClone can be adopted by others. Action research is defined as faculty-led research and dissemination of an innovation that enhances student learning (Slater et al., 2010).
Synthetic biology is a new, interdisciplinary field that blends chemistry, engineering, computer modeling, molecular biology, and systems biology. As is often the case with new fields, there is no universal definition of the field, even among practitioners. The Joint Synthetic Biology Initiative in the United Kingdom gave this definition: “Synthetic biology aims to design and engineer novel biologically-based parts, devices and systems, as well as redesign existing natural biological systems for useful purposes. It incorporates the principles of engineering (e.g., modularity, abstraction, and orthogonality) into classical biotechnology” (BBSRC, 2011, p. 2). Faculty members who are not familiar with synthetic biology might feel they cannot teach a discipline new to them. However, the heart of pClone is standard molecular biology methods incorporating four characteristics that differentiate synthetic biology from genetics or cell biology: abstraction, mathematical modeling, modularity, and standards (Knight, 2003), which are defined in the Supplemental Material.
Learning Objectives
Vision and Change (AAAS, 2011) divides its learning objectives (LOs) into two categories: core concepts and core competencies. One core concept is information flow, exchange, and storage. Promoter regulation of gene transcription falls within the core concept of information. Core competencies include the process of science, the interdisciplinary nature of modern biology, data interpretation, quantitative skills, communication in multiple formats, and experience with large databases. The pClone system helps students meet the Vision and Change expectations for the core concept centered on biological information (gene promoters) and for the six core competencies.
We used the pClone system with two different courses at our two institutions (see Student Demographics and Assessment under Methods). Biology 113 introductory biology students at Davidson College taking their first college-level course as potential majors used pClone Basic. The classroom component of Biology 113 is also consistent with Vision and Change (Barsoum et al., 2013). The pClone laboratory module emphasizes student thinking and experimental design rather than performance of predesigned experiments. After completion of the laboratory assignments and concomitant classroom activities, students who successfully complete Biology 113 will be able to meet specific LOs. They will be able to:
LO1: describe how cells can produce proteins at the right time and correct amount; | |||||
LO2: diagram how a repressor works to reduce transcription; | |||||
LO3: diagram how an activator works to increase transcription; | |||||
LO4: identify a new promoter from literature and design a method to clone it and test its function; | |||||
LO5: properly manipulate DNA and Escherichia coli for ligation and transformation experiments; | |||||
LO6: design an experiment to verify a new promoter has been cloned into destination vector; | |||||
LO7: design an experiment to measure the strength of a promoter; | |||||
LO8: distinguish type II and type IIs restriction enzymes; and | |||||
LO9: explain how Golden Gate Assembly (GGA) works. | |||||
Missouri Western State University (MWSU) second-year biology majors taking a sophomore-level, introductory genetics course used pClone Basic and pClone Green for a slightly different experimental design, as described below. Six of the nine learning objectives for genetics were identical to those for introductory biology at Davidson. Because the focus of genetics is different from introductory biology, genetics had three unique learning objectives that replaced LOs 2–4 from above. Students completing genetics will be able to:
LO2– genetics: diagram a bacterial promoter with −35 and −10 elements and the transcription start site; | |||||
LO3– genetics: describe how mutational analysis can be used to study promoter sequence requirements; and | |||||
LO4– genetics: develop a promoter mutation hypothesis and design an experiment to test it. | |||||
All of the learning objectives are compatible with the core concept of biological information and the core competencies described in Vision and Change. Our pedagogical goal was to develop an inexpensive and simple laboratory module that AP Biology and college faculty members could adopt very easily to their local conditions while accomplishing the specific learning objectives outlined above.
Cloning DNA Easily
The bane of most biologists accustomed to cloning DNA is gel purification, a method that is often dreaded by laboratory instructors. Gel purification is used to separate two or more DNA fragments so at least one fragment can be cloned into a new plasmid. The method is slow, labor-intensive, and expensive, and failure rates are high. We wanted to develop a DNA-cloning laboratory module for novices that was much easier to perform. To make cloning easy, we adapted GGA for use with the pClone vectors (Weber et al., 2011; Werner et al., 2012).
GGA relies on type IIs restriction enzymes such as BsaI (Figure 1A). Type IIs enzymes differ from the more familiar type II enzymes that bind to palindrome sequences and cut within the recognition sequences. Like all type IIs enzymes, BsaI binds to an asymmetric DNA sequence and makes a staggered, double-stranded cut in the DNA outside the recognition sequence. Because the cut is always 3′ to the binding site of 5′ GGTCTC 3′, and because this sequence can be on either the top or the bottom strand, it is useful to accompany each BsaI site label with an arrow to indicate whether the cut is to the right (GGTCTC on the top strand) or to the left (GAGACC on the top strand) of the binding site. The staggered, double-stranded cut produces a 5′ overhang sticky end, four bases in length, that is independent of the recognition sequence. The two DNA parts to be connected by GGA are engineered to have BsaI restriction sites that produce complementary sticky ends and are removed in the final ligation product. An insert is flanked by two inward-facing BsaI sites, and the sequence to be replaced in the destination vector is flanked by two outward-facing BsaI sites (Figure 1B). The insert and the destination vector are mixed with BsaI and DNA ligase simultaneously in a single tube under conditions that support both enzymes. Cutting at a BsaI site could be followed by ligation to regenerate the original DNA. However, ligation of the new insert into the vector is a terminal product, because the new insert contains no BsaI sites and therefore cannot be cut by BsaI. GGA requires type IIs enzymes and is simpler to perform than another commonly used synthetic biology method, BioBrick Assembly (Knight, 2003). The spacing between the two parts connected by GGA can be zero bases, rather than the obligatory six bases required when using BioBrick Assembly (see Supplemental Material for details). GGA is amenable to typical high school and college laboratory modules, because GGA is so easy to perform.
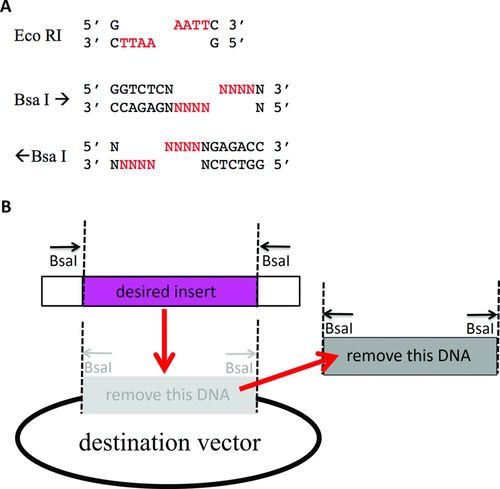
Figure 1. Type IIS restriction endonucleases enable GGA. (A) Type II enzyme EcoRI binds to its palindromic recognition sequence and makes a staggered cut resulting in a 5′ overhang that is always AATT (red). The nonpalindromic binding site of type IIs enzyme BsaI dictates the left or right direction of a staggered cut outside the recognition sequence, resulting in a 5′ overhang whose sequence is determined by the adjacent DNA (red). (B) The desired insert to be cloned with GGA is flanked by two BsaI sites that are cut (dotted lines) toward the insert, which removes the BsaI sites from the insert. The DNA to be replaced in the pClone destination vector is flanked by two BsaI sites that are cut away from the original insert to be removed and are excised as part of the old insert. Ligation of the desired insert into the vector is a dead-end product, because it contains no BsaI sites. Red arrows show the movement of DNA fragments during GGA.
In this paper, we present all the technical details that faculty members would require to evaluate how pClone works and whether they have the required equipment. To help readers decide whether the module would meet their educational goals, we provide assessment data from our students who completed the described laboratory assignments. To make sure learning gains we might measure were not the result of knowledge gained in other classes, we compared outcomes for our students with outcomes for students enrolled in comparable courses on the same campus who did not work with the pClone modules (described in Methods). We provide evidence that students on both campuses who used pClone significantly improved their understanding of a core concept and several core competencies. Given the ease of use and high degree of successful cloning of new promoters designed by students, we think other faculty members could use the pClone system to help their students realize some of the expectations described in Vision and Change (AAAS, 2011) and BIO2010 (NRC, 2003). During the laboratory sessions, students design promoters they want to test; assemble their promoters from oligonucleotides; clone their promoters into pClone plasmids; grow the plasmids in E. coli and quantify the promoters’ strength in E. coli based on reporter protein accumulation; compare their promoters with a common standard promoter; and report their results within class as well as in an open database created for student results.
METHODS
Cloning Methodologies
Most of the detailed bench work is described in the Supplemental Material and will not be addressed here. In short, students use literature and textbook information to design novel promoter sequences they want to test for functionality. Student research questions can be answered only by conducting bench research. To constrain costs and accelerate the DNA synthesis process, we limited oligonucleotides to a maximum of 70 bases, although we have used GGA to clone 4-kb inserts and tested promoters as big as 200 base pairs in pClone plasmids. Students use GGA to clone new promoters into one of the pClone plasmids and transform the plasmids into E. coli for testing. Promoter function results are recorded quantitatively and/or qualitatively. Depending on which plasmid is used, phenotypes can include green fluorescent protein (GFP), red fluorescent protein (RFP), or a blue chromoprotein (Team Uppsala-Sweden, 2011). Successful cloning of student promoters can be verified by change of colony color, polymerase chain reaction (PCR), or DNA sequencing. All of the student protocols and reagents are available in the Supplemental Material.
Measuring Promoter Strength
Students quantified RFP fluorescence for their experimental promoter, a negative control of nonfluorescent bacteria, and a positive control of bacteria expressing RFP driven by the 200–base pair Plac promoter (see part number J04450 in the Registry of Standard Biological Parts, 2013). Students measured absorbance at 590 nm for cell density to produce a relative ratio of fluorescence to absorbance. For a qualitative approach, students can generate a dilution series of experimental clones produced in pClone Blue and score the intensity of the blue color compared with positive and negative control clones (Supplemental Figure S1). Alternatively, students can measure promoter function as indicated by reporter protein abundance by using the freely available software ImageJ (Rasband, 1997–2012) to analyze photographs of pClone colonies. As described in the Supplemental Material and illustrated in Figure S2, ImageJ lets students quantify relative promoter strength of promoters cloned into all three of the pClone plasmids.
Registry of Functional Promoters
We have established an online database for student promoter results called the Registry of Functional Promoters database (Campbell et al., 2013; http://gcat.davidson.edu/RFP). Students submit their promoter sequences, relative intensities, and the experimental details, so others can compare their promoters under identical conditions. Faculty members who want to use the Registry of Functional Promoters should sign up as faculty members and direct their students to self-register as students under the name and home institution of their instructors. Faculty can edit any of the information submitted by their students, but students can edit only their own submissions. Additional details of the Registry of Functional Promoters database are available in the Supplemental Material.
Student Demographics and Assessment
Introductory Biology.
The 30 introductory biology students in the experimental group at Davidson College self-selected into this majors’ course (Biology 113) rather than into a comparison section of a similar course (Biology 111; Barsoum et al., 2013) offered during the same 2012 Fall semester. Both sections met 3 d/wk for 50 min and had a laboratory component that met 1 d/wk for 2.75 h. During registration and add/drop, none of the students were aware of the new laboratory modules using pClone. Biology 111 and 113 are the first of a two-semester series populated mostly by students interested in biology as a possible major (Barsoum et al., 2013). The comparison students (receiving a different form of instruction on promoters) had completed their genetics module in Biology 111 when they took the posttest, but they did not perform any of the pClone laboratory modules carried out by the Biology 113 students. Equivalency of the experimental and comparison students was determined using an independent t test for those students for whom SAT scores were available (n = 29 for comparison students; n = 18 for experimental students). No significant difference in total SAT scores was found (t(45) = 1.222, p = 0.228), with the comparison students having an average of 1925 (SD = 394.5) and experimental students having an average of 2045 (SD = 169.7). Although the comparison class had a slightly higher proportion of sophomores (34.4% vs. 23.3%) and juniors (6.3% vs. 0%) than the experimental class, this difference was not significant (χ2(2) = 3.209, p = 0.201). Neither class had any seniors, and both classes were dominated by first-year students.
Genetics.
MWSU students (46 Fall semester 2012; 57 Spring semester 2013) who registered for the sophomore-level genetics course for biology majors participated in the study. Control students (who received no instruction on the topic) for both semesters were taking a sophomore-level ecology course and had no instruction on the topic of transcriptional promoters. Any students who had already used the pClone module were excluded from the ecology control populations. Students registering for genetics were unaware of the new pClone laboratory modules for the Fall of 2012 and were likely to be unaware in the Spring of 2013. Students taking genetics or ecology are biology majors typically in their second year of college. We are unaware of any self-selection bias based on instructors. The only difference we know of is that those taking ecology generally do not prefer to study the molecular topics that are the subject of this intervention and assessment. We compared the students in the genetics and ecology classes based on outside measures to determine whether they showed any hidden academic differences. There was no significant difference between mean ACT composite scores for ecology students (22.76) and genetics students (22.58; t(146) = 0.270, p = 0.788). Furthermore, the number of high school core courses was indistinguishable (15.31 vs. 15.59; t(124) = 1.416, p = 0.159), and the number of college credits (13.51 vs. 13.46; t(187) = 0.073, p = 0.942) was also indistinguishable.
Assessment.
A.M.C. and T.E. wrote and graded the assessment instrument, which is available upon request to qualified instructors (the reviewers for this manuscript had access to the instruments). The instruments were administered on paper during lab periods either before the laboratory modules or 1 wk after the lab modules were completed. The comparison (Biology 111) and control students (ecology) were given the pretest instrument the same week as the experimental students took the posttest. The instruments on both campuses were identical for seven of the nine total questions. The two variable questions addressed the campus-specific learning goals as described in the Introduction.
In the Fall of 2012, students on both campuses were given identical versions of the most difficult item (LO4), which was a multiple matching question. It became clear the question had too many options, and students very rarely got the answer completely right. For the Spring of 2013, the question was simplified by eliminating several correct and incorrect choices from the long list of options. This revised item was more reasonable in length than the first iteration.
Approval of the use of human subjects for the study was obtained on both campuses with Davidson College IRB approval #2012–097 and MWSU IRB approval #2012–1591.
RESULTS
We wanted our students to achieve important learning objectives using a laboratory module that leveraged the simplicity of GGA. The core concept we wanted our students to understand was the transcriptional control of genes by bacterial promoters. Our goal was to develop a laboratory module that could provide high school and college students with access to the power of synthetic biology within the real-world constraints of typical teaching laboratories. We used the pClone plasmids in three offerings of two different courses on our campuses (see Methods). The success of using pClone in two different courses on two campuses illustrates the flexibility with which it can be used for course-specific learning objectives. At Davidson College, we used pClone Basic in an introductory biology course primarily for first-year students who might become biology majors. Before manipulating the DNA, students were asked to search the literature and find bacterial promoters that were regulated in interesting ways. Examples included promoters that were regulated by sugars, salts, amino acids, and tetracycline. In a sophomore-level genetics class for biology majors at MWSU, students were presented with the sequence of a known constitutive promoter and used it as a platform for studying the process by which transcription is initiated. The students were asked to develop and test mutational hypotheses to investigate the function of bases within known promoter elements, such as the −10 and −35 sites. Students were allowed to introduce substitution mutations (transitions or transversions), deletions, or insertions.
The first step in the project for each class was for students to determine the sequence of the promoters they wanted to clone. They used the promoter sequences as inputs into the Oligator Web tool, which generates outputs of top and bottom strand sequences with sticky ends appropriate for cloning into each of the pClone vectors (Meador et al., 2010). Oligonucleotides were ordered and received before the next lab meeting. All of these manipulations fit easily within a traditional lab period of 2.75 h (see Supplemental Material).
Characterizing Promoters in pClone Plasmids
The first destination vector we developed was pClone Basic (Figure 2A). pClone Basic contains a double transcriptional terminator between the BsaI sites used in GGA. Introduction of a functional promoter in place of the double terminator will result in transcription of a ribosome binding site (RBS) and an RFP coding sequence. The rate of mRNA production is determined by the strength of the student-built promoter. Translation of the mRNA results in RFP accumulation, which can be viewed with visible or UV light and used as an indirect measure of the transcriptional activity of the students’ promoter. In cases in which promoter activity is high enough to result in visible RFP production, the result of GGA and subsequent E. coli transformation is a mixture of red and uncolored colonies on a plate (Figure 2B, middle). Red colonies indicate successful assemblies of a functional synthetic promoter into a pClone vector. Uncolored colonies could be the result of failure to remove the transcriptional terminator (Figure 2B, left) or successful cloning of a nonfunctional promoter (Figure 2B, right). Alternatively, an uncolored colony may indicate the promoter requires additional chemical signals to initiate transcription.
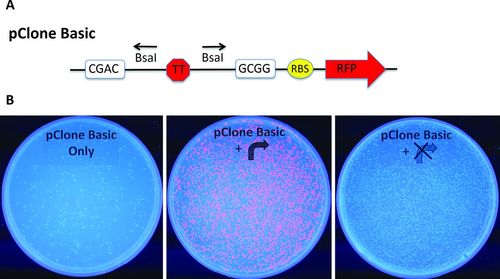
Figure 2. Cloning promoters with pClone Basic. (A) pClone Basic contains a double transcriptional terminator (TT) flanked by BsaI sites that use CGAC and GCGG sticky ends. Downstream of the BsaI sites are a ribosome binding site (RBS) and an RFP reporter gene. (B) pClone Basic negative control (left panel) produces colonies lacking RFP fluorescence under UV light. GGA with a functional promoter produces red fluorescent colonies (center panel), but GGA with a nonfunctional promoter yields colonies with the normal pale color (right panel).
Oligonucleotides can be ordered with variable bases (e.g., W represents A or T), providing students with more flexibility in their research design. Figure 2B shows the results of an experiment in which both a functional and a nonfunctional promoter were cloned into pClone Basic. The two Ptac promoter variants include mutations designed by MWSU genetics students (De Boer et al., 1983). Colonies derived from conducting GGA with pClone Basic and no promoter (negative control; Figure 2B, left) do not display fluorescence under UV light, as expected. Students also cloned a Ptac promoter that mutated the −35 control element from TTGACA to TTWACA (Figure 2B, center). Each of the red fluorescent colonies contains a mutated and functional Ptac promoter cloned into pClone Basic. Students also used GGA and pClone Basic to test a Ptac promoter that has a mutation of the −10 element from TATAAT to AAAAAA. The fact that no red fluorescent colonies are visible supports the conclusion that the mutation rendered the promoter nonfunctional. However, in the experiments shown in both the center and the right portions of Figure 2B, colonies containing the original pClone plasmid with the transcriptional terminator are indistinguishable from colonies containing a new, nonfunctional promoter. Through DNA sequencing and PCR amplification, we have verified promoter cloning leading to weak expression of protein (see Supplemental Material and Figure S3).
As a result of questions our students raised about their in-class experiments using pClone Basic, two of the authors (T.E. and A.M.C.) designed and built two new pClone plasmids called pClone Green (Figure 3A) and pClone Blue (Figure 4A) that distinguish plasmids lacking a new promoter from plasmids containing new but nonfunctional promoters. The two new plasmids eliminate the need for time-consuming screens to identify plasmids that contain new, student-generated promoters. Figure 3B shows the results of using pClone Green to clone the same two Ptac mutant promoters studied in Figure 2B. Only green fluorescent colonies are visible in Figure 3B (left), because these cells received DNA from the negative control ligation, which lacked any new promoter DNA. Figure 3B (center) shows the results of GGA with the TTWACA mutant promoter and pClone Green. In this experiment, green colonies are produced by the original pClone Green plasmid. Close examination of the plate reveals both red and uncolored colonies. Subsequent DNA sequencing verified that the red colonies are the result of cloning the TTWACA mutation with W = T, while the uncolored colonies have W = A. GGA with the −10 AAAAAA mutant promoter and pClone Green is shown in Figure 3B (right). Green fluorescent colonies contain the original pClone Green plasmid. Colonies that do not fluoresce contain a nonfunctional mutant promoter.
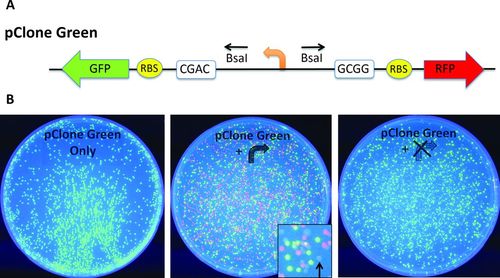
Figure 3. Cloning promoters with pClone Green. (A) pClone Green contains a promoter in reverse orientation as a starting insert between BsaI sites that use CGAC and GCGG sticky ends. The promoter initiates transcription of a GFP gene in reverse orientation preceded by an RBS. Insertion of a functional promoter will lead to transcription of the RFP gene preceded by an RBS. (B) pClone Green negative control produces colonies that display GFP fluorescence under UV light (left panel). GGA with a functional promoter yields a mixture of green fluorescent colonies containing the starting pClone Green promoter, red fluorescent colonies containing a strong functional promoter, and nonfluorescent colonies (magnified inset, black arrow) with no promoter (center panel). GGA with a nonfunctional or inactive promoter yields a mixture of green fluorescent colonies containing the starting pClone Green plasmid and nonfluorescent colonies containing the nonfunctional or inactive promoter (right panel).
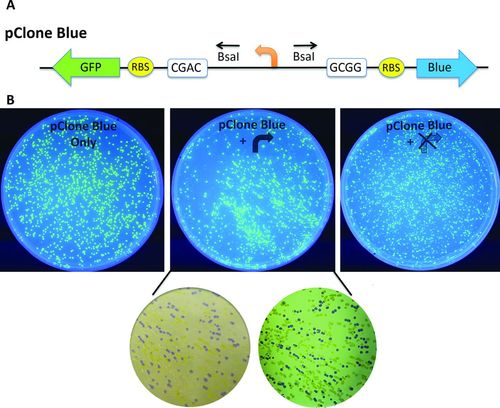
Figure 4. Cloning promoters with pClone Blue. (A) pClone Blue contains a promoter in reverse orientation between BsaI sites that use CGAC and GCGG sticky ends. The promoter drives transcription of a GFP gene in reverse orientation preceded by an RBS. Insertion of a functional promoter will lead to transcription of the AmilCP Blue gene preceded by an RBS. (B) pClone Blue negative control produces colonies that display GFP fluorescence under UV light (left). GGA with a functional promoter (center) yields a mixture of green fluorescent colonies containing the starting pClone Green plasmid and colonies that are blue with either overhead white light (bottom left) or on a white light box (bottom right). GGA with a nonfunctional or inactive promoter yields a mixture of green fluorescent colonies containing the starting pClone Green plasmid and nonfluorescent, nonblue colonies containing the nonfunctional or inactive promoter (right).
Knowing that many schools do not have fluorometers to quantify RFP production, we designed pClone Blue, which uses chromoprotein AmilCP Blue (Figure 4A; Team Uppsala-Sweden, 2011)). Like pClone Green, pClone Blue includes a reverse-orientation GFP gene transcribed by a reverse-orientation promoter flanked by two BsaI sites used for GGA. Failure to clone a new promoter results in colonies that express GFP. Successful assemblies result in blue colonies, if the synthetic promoter is active, or uncolored colonies, if the promoter is not active. The student-designed TTWACA mutant promoter and pClone Blue produced green fluorescent colonies that are visible under UV light (Figure 4B, center) resulting from the starting plasmid. Colonies that contain the functional TTWACA mutation with W = T are blue when visualized with overhead white light or on a white-light box (Figure 4B, center). Uncolored colonies derived from the nonfunctional mutant promoter with W = A can also be observed on the plate. Using GGA and the nonfunctional −10 AAAAAA mutant promoter with pClone Blue produced green fluorescent colonies containing the original left-facing pClone Blue promoter and nonfluorescent colonies containing the mutant promoter (Figure 4B, right).
Quantifying Promoter Strength
Plated colonies, such as those shown in Figures 2–4, can indicate whether promoters are functional or not as a result of mutation or because of regulation. The pClone plasmids can also be used to quantify promoter function. The intensity of red fluorescence resulting from RFP expression in pClone Basic or pClone Green experiments can be used as a measure of the relative strength of a given promoter. RFP reporter gene expression is best measured with a fluorometer. Figure 5 shows student data of a promoter that did not function the way students had hypothesized. Introductory biology students at Davidson used GGA to ligate a version of the E. coli cydAB promoter into pClone Basic. Students grew the engineered bacteria in liquid culture overnight in the presence or absence of a predicted inducer. They expected to detect increased transcription. Instead, they found that their uninduced promoters were 67 and 78% as strong as the Plac positive-control promoter. The data also show that the level of transcription from their promoter decreased with exposure to the predicted inducer. These results were unexpected and raised many questions for the students to address in their lab reports and oral presentations.
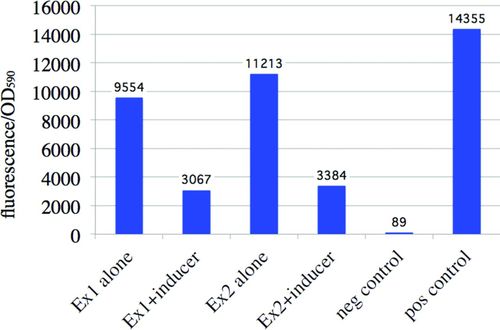
Figure 5. Measurement of promoter activity using the pClone system. Student-built promoter (J100099) cloned into pClone Basic. The ratio of RFP fluorescence at 615 nm is divided by absorbance at 590 nm as a measure of cell density. This figure was generated by undergraduates who tested two clones of the same promoter (Ex1 and Ex2), a negative control of pClone Basic with the transcriptional terminator, and a positive control promoter in part J04450. Students predicted that the inducer (NH4)2Fe(SO4)2 would stimulate the promoter, but their hypothesis was incorrect.
Another method to quantify RFP or AmilCP Blue chromoprotein expression is to use the freely available ImageJ program (Rasband, 1997–2012) to analyze photographs of plates with pClone colonies. We used ImageJ to analyze the center plate images in Figures 2B, 3B, and 4B. As described earlier, these plates each contain two cloned promoter mutations, one that resulted in a strong promoter and one that resulted in a weak promoter. The ImageJ procedure (see Methods) generated expression ratios comparing the strong (W = T in TTWACA) with the weak (W = A in TTWACA) promoter for four colonies of each type on all three plates. The expression ratio was 2.13 for the pClone Basic plate, 2.20 for the pClone Green plate, and 2.21 for the pClone Blue plate (see Supplemental Material and Figure S2). The expression of AmilCP Blue in the pClone Blue plasmid can also be assessed visually or with a spectrophotometer (see Supplemental Material and Figure S1).
Sharing Results with Research Community
Students practiced the core competency of communicating science by presenting their results. Students prepared oral and written presentations on their experimental design and results. They also registered the new synthetic promoter clones they had produced in the Registry of Standard Biological Parts (2013), thereby sharing their results with the global synthetic biology research community (Campbell, 2005). Students also entered their results into the Registry of Functional Promoters database (Campbell et al., 2013), which is a new database of student-designed and tested synthetic promoters. The Registry of Functional Promoters database prompts students to include the necessary information that will allow others to directly compare multiple promoters. This comparison of promoters is what will make the undergraduate-generated database a powerful source of information for synthetic biology researchers. This open-access database converts undergraduate lab projects into meaningful research results, because the data are publicly available.
Measuring Learning Gains
Typically, action research does not compare experimental student learning gains with assessment results of other students. The purpose of comparing our experimental students with other students was to help determine whether students in a similar class who did not work with pClone plasmids could score equally well on our assessment instruments. We considered that any learning gains we measured could be due to general improvements in the overall college experiences of the students. One way to address this concern is to use previously published assessment instruments (Klymkowsky et al., 2003; Bowling et al., 2008; Smith et al., 2008), but none of those instruments are aligned with our specific learning objectives. Therefore, it was important for us to determine how comparison (introductory biology students in different sections) and control (ecology) students performed at the same point in the semester as our experimental students. The comparison introductory biology class shared only LO1 (to understand the role of a promoter). As would be expected, the ecology course did not share any of the genetics learning objectives. Control and comparison students of similar academic characteristics took their test the same week the experimental class took their posttest.
Using a pretest and a posttest, we measured significant learning gains for students who used the pClone system at Davidson College and MWSU. The questions on the pretest and posttest combination were identical for a given group of students. The questions were designed to assess the specific learning objectives (listed in the Introduction) related to bacterial gene expression and regulation and to molecular cloning by GGA. We assessed Fall 2012 introductory biology students at Davidson, Fall 2012 genetics students at MWSU, and Spring 2013 genetics students at MWSU.
At Davidson College, 30 students enrolled in introductory biology used the pClone laboratory module and were assessed in each of nine learning objectives at the start of the course and again after completing an oral presentation of their pClone research results. Thirty-one students in a comparison section of introductory biology at Davidson College did not use the pClone system and were assessed for the nine learning objectives at the same time in the semester as the experimental group took their posttest. Differences in learning objectives between the three sets of scores (pretest, posttest, comparison) were assessed using a one-way analysis of variance (ANOVA) for each of the nine learning objectives. Where there were significant differences, Tukey's honestly significant difference (HSD) was used to determine which groups of students had different test results (Table 1). The experimental students showed significant improvement in LOs 1, 2, 3, 4, 5, and 9 compared with both their own pretest scores and the scores of students in the comparison introductory biology course, which did not incorporate the use of pClone. The test scores of the comparison group and the pretest scores of the experimental group were equivalent. Furthermore, the effect sizes for most of these differences were large, indicating that pClone is not only effective but has a substantial impact on improving scores in these six learning objectives (Maher et al., 2013)
| Learning objective | Pretest experimental | Posttest experimental | Comparison course | F(2,88) | Effect size (η2) | Conclusion | |
|---|---|---|---|---|---|---|---|
| 1 | Function of promoter | 43% | 87%a | 48% | 8.008, p = 0.001 | 0.154 | Large effect |
| 2 | Repressor diagram | 23% | 53%a | 13% | 7.206, p = 0.001 | 0.141 | Large effect |
| 3 | Activator diagram | 0% | 41%a | 0% | 7.250, p = 0.001 | 0.167 | Large effect |
| 4 | Experiment overview | 0% | 13%a | 0% | 4.538, p = 0.013 | 0.103 | Moderate effect |
| 5 | Transformation method | 0% | 20%a | 0% | 7.374, p = 0.001 | 0.143 | Large effect |
| 6 | Verify promoter cloned | 50% | 40% | 48% | 0.34, p = 0.713 | 0.008 | No effect |
| 7 | Test promoter strength | 43% | 60% | 39% | 1.525, p = 0.223 | 0.034 | No effect |
| 8 | Type IIs restriction enzymes | 7% | 50% | 6% | 1.873, p = 0.16 | 0.041 | No effect |
| 9 | GGA method | 10% | 63%a | 0% | 31.929, p < 0.001 | 0.421 | Large effect |
Our data indicated that pClone was effective at helping students achieve the learning objectives in an introductory biology course at Davidson College, a highly selective liberal arts institution. We wanted to know whether similar gains could be detected in a sophomore-level course (genetics) at MWSU, a comprehensive regional university with open admissions. We used a sophomore-level control course (ecology) to assess whether similar biology majors at MWSU could score equally well on the assessment instrument. Differences in scores between the three sets of scores (pretest, posttest, ecology control) were assessed using a one-way ANOVA for each of the nine learning objectives. Where there were significant differences, Tukey's HSD was used to determine which groups of students had different test results (Table 2). The genetics students showed significant gains in LOs 1, 2, 3, 5, 6, 7, and 8. Their scores on these questions were significantly better than the ecology students’ scores. The genetics pretest scores and ecology scores were equivalent to each other. Furthermore, the effect sizes for most of these differences were large or moderate, indicating that pClone is not only effective, but it has a meaningful impact on improving scores in the seven learning objectives.
| Learning objective | Pretest experimental | Posttest experimental | Control course (ecology) | F(2252) | Effect size (η2) | Conclusion | |
|---|---|---|---|---|---|---|---|
| 1 | Function of promoter | 36% | 59%a | 20% | 13.527, p < 0.001 | 0.097 | Moderate effect |
| 2 | −10 and −35 sites | 3% | 70%a | 0% | 145.374, p < 0.001 | 0.536 | Large effect |
| 3 | Mutational analysis | 30% | 75%a | 33% | 28.773, p < 0.001 | 0.186 | Large effect |
| 4 | Student-designed mutation | 0% | 0% | 0% | 0, p > 0.05 | 0.000 | No effect |
| 5 | Transformation method | 11% | 51%a | 12% | 30.731, p < 0.001 | 0.196 | Large effect |
| 6 | Verify promoter cloned | 19% | 44%a | 18% | 10.264, p < 0.001 | 0.075 | Moderate effect |
| 7 | Test promoter strength | 17% | 33%a | 18% | 4.421, p = 0.013 | 0.034 | Moderate effect |
| 8 | Type IIs restriction enzymes | 2% | 29%a | 4% | 21.661, p < 0.001 | 0.147 | Large effect |
| 9 | GGA method | 14% | 22% | 14% | 1.56, p = 0.212 | 0.012 | No effect |
DISCUSSION
This paper describes action research with assessment data that document the effectiveness of the educational innovation (Slater et al., 2010). In publishing our work, we hope that other faculty will use the pClone suite of plasmids to improve student learning in ways called for in Vision and Change and the AP Biology redesign (College Board, 2011; Wood, 2002; Barsoum et al., 2013). We are aware that action research typically does not have control or comparison groups, but we wanted to determine whether comparable, nonexperimental students could score equally well on our assessment. We did not use previously published assessment instruments (Klymkowsky et al., 2003; Bowling et al., 2008; Smith et al., 2008), because none of them is aligned with our specific learning objectives (listed in the Introduction).
The pClone family of three plasmids provides an inexpensive entry point to synthetic biology and promoter analysis that faculty members can employ to offer authentic research experiences for their students within the context of laboratory classes. pClone leverages the modern method of GGA (Weber et al., 2011; Werner et al., 2012) as a cutting-edge molecular cloning tool that can be used by students with little or no experience in molecular biology. Our experiences with pClone in introductory biology and sophomore-level genetics demonstrated that students successfully cloned and tested novel bacterial promoters. Students were eager to accept the challenge of designing their own promoters and were able to quickly learn the laboratory skills needed to perform their experiments. Our students commented that they appreciated the opportunity to engage in genuine research within the context of a laboratory class. The way we used pClone in our classes would translate well to a variety of institutional settings. All of the common barriers to molecular cloning experiments of cost, faculty expertise, equipment, and limited class time are minimized by the pClone system.
Both courses shared six of the nine LOs. For LO1 (function of promoter) and LO5 (transformational method), the pClone module was effective on both campuses (Table 3). For three learning objectives (LO6: verify promoter cloned; LO7: test promoter strength; and LO8: type IIs restriction enzymes) the pClone experience was effective for sophomores in genetics but not for introductory biology students. For LO9 (GGA method), the pClone module was effective for introductory biology students but not for students enrolled in genetics. It is unclear why LOs 6–9 have campus-specific differences, but perhaps the differences are related to the amount of time spent on topics during lab. The introductory lab module spans 8 wk, and the students are conducting a second laboratory module concurrently during the last 4 wk of the pClone module. Genetics, on the other hand, spends 3 wk on this module while concurrently conducting other experiments. We plan to reevaluate the importance of our original nine LOs and determine whether or not they are important enough for us to invest more time to address them. For example, LO6 (verify promoter cloned) is no longer important, because pClone Green and pClone Blue make the objective obsolete.
| Learning objective | Davidson introductory biologya | MWSU geneticsa | Impact on students | |
|---|---|---|---|---|
| 1 | Function of promoter | Large effect | Moderate effect | Universally effective |
| 2I | Repressor diagram | Large effect | n. a. | Effective for intended students |
| 2G | −10 and −35 sites | n. a. | Large effect | Effective for intended students |
| 3I | Activator diagram | Large effect | n. a. | Effective for intended students |
| 3G | Mutational analysis | n. a. | Large effect | Effective for intended students |
| 4I | Experiment overview | Moderate effect | n. a. | Modestly effective for intended students |
| 4G | Student-designed mutation | n. a. | No effect | Modestly effective for intended students |
| 5 | Transformation method | Large effect | Large effect | Universally effective |
| 6 | Verify promoter cloned | No effect | Moderate effect | Effective for more advanced students |
| 7 | Test promoter strength | No effect | Moderate effect | Effective for more advanced students |
| 8 | Type IIs restriction enzymes | No effect | Large effect | Effective for more advanced students |
| 9 | GGA method | Large effect | No effect | Effective for introductory students |
In Figure 5, students graphed their fluorescence data divided by OD600. It is important that students realize the need to divide the amount of RFP fluorescence by the density of cells (OD600). Generating this ratio is a good example of how students learn core competencies of data interpretation and quantitative skills. It is easy for students to intuit that cultures with twice as many cells should appear twice as colorful as another culture. The pClone laboratory module focuses on the core concept of transcription and gene regulation rather than cell density. What we really wanted to study was the promoter strength as measured by amount of fluorescence for a given cell density. By dividing by the density of cells, students essentially collect a colorimetric reading of transcriptional output per cell.
By reporting the relative strength of their cloned promoters in E. coli, students enable synthetic biologists from anywhere in the world to indirectly compare the strength of student promoters, even though the promoters were not tested simultaneously. Because fluorescence is measured in arbitrary units, it is not appropriate to compare numerical values across experiments. For this reason, students reporting the function of new promoters should compare their promoters with the universal standard promoter Plac (see part number J04450). In the Registry of Functional Promoters database developed for use with pClone plasmids, introductory biology students reported the strength of their promoters relative to the promoters’ strength in part number J04450. Students reporting the effect of mutations on a promoter should report expression ratios that reflect the relative strength of the mutants in the data set. Genetics students also reported the results for their mutation experiments involving Ptac in the Registry of Functional Promoters database. This database contains information pertaining to the growth conditions, strain of E. coli, and other variables that will facilitate meaningful comparisons between student-tested promoters. All of this important comparative information will make it easy for synthetic biology researchers to choose promoters that will function appropriately for their synthetic biology projects.
Students using pClone plasmids are conducting real research on bacterial transcriptional promoters. Introductory biology students who conducted classroom research can feel empowered and might develop the confidence they need to pursue independent undergraduate research, either in synthetic biology or in another area of biology (Harrison et al., 2011). Several of the introductory biology students from this class have pursued independent study research after they used pClone. Two of our genetics students using pClone Basic engaged in an independent research project of constructing pClone Green and pClone Blue, thereby earning authorship on this publication (C.S. and P.F.). Synthetic biology and the pClone system are also accessible to high school students. This is exemplified by three high school researchers who constructed and tested pClone Basic and another who developed the pClone Blue measurement protocol. All four of the high school students (C.A., S.H., A.W., and J.Y.) earned authorship on this publication based on research they conducted with pClone.
The pClone plasmids could serve as a platform for a variety of independent research projects. Students could use pClone to further investigate the regulation of a promoter they had cloned in class, exploring the types and concentrations of inducers or corepressors needed to effect changes in the level of transcription. A given promoter could be mutated at various positions to provide information on its function. Students could take an engineering approach to the production of hybrid promoters or promoters that respond to new chemical inputs. For example, Figure 5 shows students incorrectly predicted (NH4)2Fe(SO4)2 would stimulate their promoter, but it repressed the promoter instead. Unexpected results such as these could be the starting place for additional research. Students could explore the significance of plasmid copy number by moving their constructs to a plasmid with a different origin of replication. These and other projects become feasible for undergraduates and high school students with the pClone system.
The motivation for this project was centered on student learning. As recommended by BIO2010 (NRC, 2003) and Vision and Change (American Association for the Advancement of Science, 2011), we have developed a laboratory module that accomplishes several important objectives. BIO2010 emphasized the importance of authentic research using modern methods. Synthetic biology is a new discipline that blends molecular biology methods with the quantitative disciplines of mathematics and engineering (Knight, 2003). With pClone, students think of functional DNA parts as modular bits that can be interchanged easily. Students also must consider standards of measurement, which facilitates comparison of promoter function. As pointed out in numerous studies, early research experiences can have multiple benefits for a diverse population of students considering science, technology, engineering, and mathematics majors (Seymour et al., 2004; Lopatto, 2006; Laursen et al., 2010).
Vision and Change encouraged curriculum reform that addressed core concepts and competencies. One of the core concepts is information, including information on gene regulation and transcription. Core competencies include the process of science, interdisciplinary nature of modern biology, data interpretation, quantitative skills, communication in multiple formats, and student experience with large databases. The pClone plasmids allowed our students to gain firsthand experience in each of these competencies. It is very rare that a single laboratory module can address so many important areas of learning. By presenting their research results to their peers, the students gained practice in oral and poster communication formats. Many students used GenBank and other tools from the National Center for Biotechnology Information as part of their promoter literature searches. In addition to being consumers of databases, our students contributed content to the databases of Registry of Standard Biological Parts (2013) and Registry of Functional Promoters. It is unusual for undergraduates to be authors in research-grade databases. The pClone system allows genuine student research to become commonplace.
Based on assessment results described in this study, our students learned important concepts related to gene regulation in bacteria (Table 3). When conducting education research, it is common to hear skeptics say that the investigators simply taught to the test and that is why the scores improved. However, when the learning objectives are established first and the assessment instrument is in alignment with the desired outcomes, teaching to the test is what we should all do. The comparison and control students were the same age as and had a level of instruction similar to our two populations of experimental students. Our experimental students could not complete learning objectives at the beginning of the class, but our students knew significantly more than the comparison or control students later in the semester. Our students learned about gene regulation, the function of promoters, and many laboratory skills associated with real research. In the process, they produced authentic research results that will be publicly accessible for many years. Through a distributed network of undergraduates working at their home institutions, faculty can use pClone and crowdsourcing to build a Registry of Functional Promoters that can be accessed by anyone looking for a promoter with defined characteristics.
ACKNOWLEDGMENTS
We thank the students of the Fall 2012 Biology 113 class at Davidson College and the Fall 2013 and Spring 2013 Biology 205 classes at MWSU for test-driving the pClone system and participating in educational research. Thanks to the Fall 2013 Biology 205 class at MWSU for designing and cloning the promoters in Supplemental Figure S2. We also thank Drs. Mark Barsoum and Dave Ashley for granting us access to their students for use as comparisons and Dr. Steve Hatch for the suggestion of using ImageJ. Support is gratefully acknowledged for National Science Foundation RUI grants MCB-1120578 to Davidson College and MCB-1120558 to MWSU; Howard Hughes Medical Institute grants 52005120 and 52006292 to Davidson College; the Davidson Research Institute and James G. Martin Genomics Program at Davidson College; and the MWSU Program of Research, Teaching, and Applied Learning (PORTAL). A.M.C. and T. E. are members of GCAT SynBio.



