A Simple and Effective Protein Folding Activity Suitable for Large Lectures
Abstract
This article describes a simple and inexpensive hands-on simulation of protein folding suitable for use in large lecture classes. This activity uses a minimum of parts, tools, and skill to simulate some of the fundamental principles of protein folding. The major concepts targeted are that proteins begin as linear polypeptides and fold to three-dimensional structures, noncovalent interactions drive this folding process, and the final folded shape of a protein depends on its amino acid sequence. At the start of the activity, students are given pieces of insulated wire from which they each construct and fold their own polypeptide. This activity was evaluated in three ways. A random sample of student-generated polypeptides collected after the activity shows that most students were able to create an appropriate structure. After this activity, students (n = 154) completed an open-ended survey. Their responses showed that more than three-quarters of the students learned one or more of the core concepts being demonstrated. Finally, a follow-up survey was conducted seven weeks after the activity; responses to this survey (n = 63) showed that a similar fraction of students still retained these key concepts. This activity should be useful in large introductory-level college biology or biochemistry lectures.
INTRODUCTION
Gaining an understanding of the fundamental principles of protein folding is an important part of most introductory-level undergraduate biology and biochemistry courses. At the introductory level, an appropriate understanding of protein folding would include the following three major concepts:
A protein is synthesized as a linear chain of amino acids that then folds into a complex three-dimensional shape.
Noncovalent interactions (hydrogen bonds, electrostatic interactions, van der Waals bonds, and hydrophobic interactions) as well as covalent interactions (e.g., disulfide bonds) between different parts of the protein chain drive the folding process.
The final folded shape of the protein depends on its amino acid sequence.
As an example of protein folding, I have used molecular visualization presentation of hemoglobin in my lectures for several years (White et al., 2002). During this presentation, I would explain how the protein began as a linear chain and then folded, citing key noncovalent interactions that give the protein its final folded shape. Although I have been pleased with the results, I wanted to find a hands-on physical model that would show how the folding process proceeds rather than working backward from a fully folded protein. I wanted each of the roughly 200 students in lecture to simulate the folding of their own protein so as to get a visual and tactile “feel” for the process. I wanted this activity to show how a linear chain of amino acids can fold into a three-dimensional shape based on interactions between parts of the chain even if the activity did not simulate all of the factors involved in this process. Furthermore, I wanted this activity to fit within the spatial, temporal, and logistic constraints of a 50-min lecture in a typical large lecture hall. Consequently, the activity needed to be inexpensive, require a small number of parts, and require no special tools or accessories.
Research on the efficacy of hands-on modeling of chemical and biological structures has shown encouraging results. There have been several studies (Howe and Durr, 1982; Copolo and Hounshell, 1995; Barnea and Dori, 1999) where use of physical models of simple chemical structures, combined with other innovations, showed increased learning compared with traditional curricula. Gabel and Sherwood (1980) showed that a year-long, model-based chemistry curriculum had improved learning outcomes compared with traditional instruction. Roberts et al. (2005) showed that physical models combined with MolVis helped students to learn about protein structure. In their study, students reported that hands-on modeling using rigid models of fully folded proteins was the most helpful mode for teaching these concepts. These results, combined with my experience teaching this material, suggested that using a hands-on activity in lecture would likely be productive.
Several hands-on protein folding activities have been developed by others. Martz (2005) recommends using Toobers—long wires wrapped in foam rubber—to simulate protein folding. Others have developed a set of highly realistic activities using Toobers with attached magnetic side chains (www.moleculardesigns.com/toobers.php). Unfortunately, although Toobers are relatively inexpensive, it is still prohibitively costly to give each of the 200+ students in lecture a Toober kit of their own. Nelson and Goetze (2004) developed an elegant demonstration of protein folding that uses pipe cleaners to simulate the protein chain. However, distributing and assembling the several pipe cleaners, tape, and binder clips required for this activity would not be easy or rapid on the small desks found in most large lecture halls.
Given these constraints, I have developed a simple simulation that uses a single piece of wire to simulate a small polypeptide chain. This activity illustrates the three core concepts of protein folding described and can be easily carried out by students in a large lecture class. I have conducted a preliminary evaluation of the activity that shows that it is effective in conveying these concepts.
INSTRUCTIONAL CONTEXT
I use this activity in General Biology I (Bio 111) at University of Massachusetts (Boston, MA). Bio 111 is the first course of a two-semester introductory biology series for biology majors. Bio 111 covers genetics, biochemistry, cell biology, and molecular biology. The course consists of a 50-min lecture three times a week and 10 laboratory sections that meet once a week for 3 h. I give the lectures and supervise the graduate teaching assistants who teach the laboratories.
There are roughly 240 students enrolled in the class in any given semester. The students in Bio 111 are a rather diverse group compared with many introductory-level science courses; about 75% are female, 40% are nonwhite, and their average age is 22 yr. Roughly one-third (29%) are majors in biology-related subjects (biology, biochemistry, premedical), and a similar fraction (35%) are undecided as to their major. For many of these students, 48% of whom are freshmen, this course is their first college science course and their first science course in several years.
I am able to determine the number of students attending each lecture by using the Personal Response System (www.gtcocalcomp.com/interwriteprs.htm), where each student answers multiple-choice questions in lecture by using an individual infrared transmitter. On the day that I conducted the first phase of the evaluation of this activity, there were 189 students in the lecture hall.
I use this activity during my first lecture on protein structure. Before this lecture, I have introduced basic chemistry as it applies to proteins (covalent and noncovalent bonds) and the idea of polymers. I begin this lecture by talking about amino acid structure—backbone and side chain—and then describe how amino acids are linked to form protein chains. I then discuss, with examples, the different properties of amino acid side chains. Finally, I show them the three-dimensional shape of hemoglobin by using molecular visualization software and explain that proteins have complex three-dimensional folded shapes. This sets up the background for the activity.
LECTURE ACTIVITY
For the activity, each student is provided with a 4-ft length of 18-gauge insulated wire. This wire is available from many sources.1 In addition, each student receives a handout that explains how to use the wire to simulate a polypeptide chain. This handout can be accessed online in .pdf and .doc format at http://intro.bio.umb.edu/BW/WireDemo.html. These instructions, combined with guidance from the lecturer, are sufficient for the students to perform the activity during the lecture.
First, I explain how their wire will model a short polypeptide chain (as shown in Figure 1). The straight part of the wire simulates the backbone, and the loops simulate the side chains. Large open loops (large enough to accommodate two fingers) represent hydrophobic side chains, long closed loops (four twists) represent positively charged side chains, and short closed loops (two twists) represent negatively charged side chains (Figure 2).
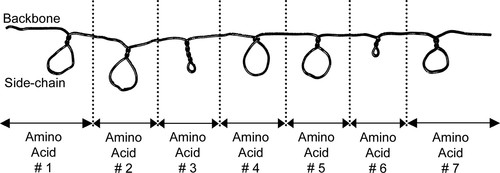
Figure 1. Model unfolded protein chain from the instruction sheet.
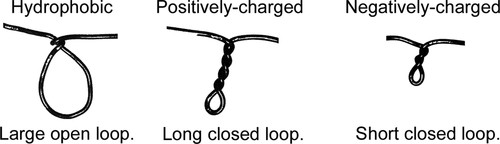
Figure 2. Correspondence between different wire loops and the side chains they represent.
Students are then told to twist their wire to make their own six- to eight-amino acid polypeptide chain. They twist the wire to form loops corresponding to their choices of hydrophobic, anionic, or cationic side chains, leaving three finger-widths of space between each side chain. The simulation works better if they make more hydrophobic side chains than charged side chains and if they also make at least one anionic and one cationic side chain. It takes students ∼3–4 min to twist their wires to produce their linear polypeptides.
At this point, I tell the students that they have just made a simulated short polypeptide. I ask them what they would expect the hydrophobic side chains to do when surrounded by water. Based on our discussions of the hydrophobic effect, they predict that the hydrophobic side chains would cluster in the center of the protein. I tell them to do this with their wires; they then fold their wires into compact three-dimensional structures with the hydrophobic side chains inside. I then ask them what the anionic and cationic side chains would do. When they respond with “form ionic bonds,” I tell them to do this with their models as well; they bring the anions to the cations as much as possible while keeping the hydrophobic side chains inside.
At this point, they each have a little ball of wire simulating their fully folded protein. I then ask them the following questions and discuss the answers (in parentheses):
What holds your polypeptide in its shape? (Ionic bonds and hydrophobic interactions.)
Why is the shape of your polypeptide different from that of your neighbor? (They have different amino acid sequences.)
EVALUATION
The first part of the evaluation was designed to assess the degree to which they understood and followed the directions. I collected a sample of 24 folded wires after the lecture. The majority (19 or 79%) had between six and eight amino acids. Twelve of the 24 “polypeptides” (50%) made by the students had a mixture of hydrophobic, cationic, and anionic side chains; another eight (33%) had hydrophobic and either cationic or anionic side chains. Only four of the 24 (17%) polypeptides had only hydrophobic side chains or were uninterpretable. Overall, 79% had an acceptable structure: six to eight side chains and a mixture of hydrophobic and charged side chains. Several wires from this sample are shown in Figure 3. The best of these (Figure 3a) clearly shows the different side chain types; others (Figure 3, b–d) show some of the mistakes that students made. These mistakes include having more hydrophilic than hydrophobic side chains (Figure 3b), having only hydrophobic and cationic side chains (Figure 3c), and not twisting the wire properly (Figure 3d).
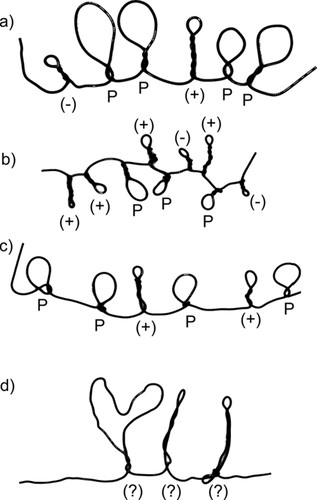
Figure 3. Some sample unfolded protein chains made by students. Side chain types are indicated as follows: P, hydrophobic; +, cationic; and −, anionic. Wires a, b, and c are satisfactory; wire d is not.
The second part of the evaluation assessed their learning of the three major concepts by using a short anonymous voluntary survey conducted in lecture immediately after the activity. A voluntary survey was chosen for its simplicity of administration; furthermore, response rates to previous voluntary surveys have been high. There were two forms of the survey distributed randomly to the class. These two different surveys were used to approach the students' learning from different perspectives and thereby yield a richer set of responses. Roughly one-half of the students received survey A, which asked the following:
What did you learn about proteins from the wire demonstration?
What about proteins was still unclear after the wire demonstration was finished?
How could the wire demonstration be improved?
Give one feature of the wire demonstration that accurately simulates the way proteins fold and explain why it is realistic.
Give a feature of the wire demonstration that does not accurately simulate the way proteins fold and explain why it is unrealistic.
How could the wire demonstration be improved?
When asked what they had learned from the demonstration (survey A, question 1), 63 responses (77%) mentioned at least one of the major concepts. When asked what was still unclear (survey A, question 2), only 11 (13%) of the responses listed any of the major learning goals. Similarly, 59 (82%) of the responses reported that at least one of the major concepts was represented accurately by the activity (survey B, question 1) and only 11 (15%) of the responses indicated that one of these was not represented accurately by the activity (survey B, question 2).
These results can be further analyzed by the specific type of response, as shown in Tables 1 and 2. Table 1 shows the eight most frequent response classes to the questions that asked for what the students learned or what was represented accurately (survey A and B, question 1). The three major concepts are the most frequently given responses. Students' responses also show several other notable learning outcomes including a “feel” for the process and an understanding of the difference between backbone and side chains. Table 2 shows the eight most frequent response classes to the questions that asked for what was still unclear or not accurately represented by the activity (survey A and B, question 2). Only one of the three learning goals (Concept [2]) is represented in these responses and was only mentioned by three students (4%). The most frequent category of response was that the exercise was fine as it is. These responses also reveal some physical difficulties with the wire and suggest that taking more time with the activity would be productive.
| What did you learn about proteins from the wire demonstration? (n = 82) | Give one feature of the wire demonstration that accurately simulates the way proteins fold and explain why it is realistic. (n = 72) |
|---|---|
| 1. 35 (43%) Hydrophobic/hydrophilic interactions. Concept (2). | 1. 45 (62%) Hydrophobic/hydrophilic interactions. Concept (2). |
| 2. 32 (39%) Protein folding process. Concept (1). | 2. 20 (28%) Protein folding process. Concept (1). |
| 3. 19 (23%) Ionic interactions. Concept (2). | 3. 11 (15%) Ionic interactions. Concept (2). |
| 4. 11 (13%) Sequence determines structure. Concept (3). | 4. 5 (7%) Backbone versus side chain. |
| 5. 11 (13%) Denaturation. | 5. 5 (7%) A tactile or visual ′feel′ for what was going on. |
| 6. 10 (12%) Vague mention of ′proteins′ only. | 6. 4 (6%) A protein as a three-dimensional object. |
| 7. 8 (10%) A tactile or visual ′feel′ for what was going on. | 7. 3 (4%) Denaturation. |
| 8. 7 (8%) Miscellaneous. | 8. 3 (4%) Vague mention of ′proteins′ only. |
| What about proteins was still unclear after the wire demonstration was finished? (n = 82) | Give a feature of the wire demonstration that does not accurately simulate the way proteins fold and explain why it is unrealistic. (n = 72) |
|---|---|
| 1. 50 (61%) ′Nothing,′ blank, ′all clear.′ | Biological issues |
| 2. 14 (17%) Questions beyond the scope of the demonstration. | 1. 20 (28%) ′Nothing,′ blank, ′fine,′ etc. |
| 3. 4 (5%) Denaturation. | 2. 12 (17%) ′In real life, a protein only folds one way.′ |
| 4. 3 (4%) ′How the amino acids are joined by the backbone.′ | 3. 6 (8%) ′Wire does not show how interactions work.′ |
| 5. 3 (4%) Ionic or hydrophobic/hydrophilic interactions. Concept (2). | 4. 6 (8%) Issues beyond the scope of the demonstration. |
| 6. 2 (2%) ′I’m not sure—I’m just a little fuzzy.′ | 5. 5 (7%) ′Real proteins have more amino acids.′ |
| 7. 2 (2%) Backbone versus side chain. | 6. 4 (6%) ′Folding wire is not like real folding.′ |
| 8. 2 (2%) Uninterpretable. | 7. 4 (6%) ′It's wire.′ |
| 8. 3 (4%) ′Wire does not show structure completely.′ | |
| Physical/logistical issues | |
| 1. 4 (6%) ′Hard to fold the wire.′ | |
| 2. 3 (4%) ′Wire too floppy.′ | |
| 3. 2 (3%) ′Took too short a time.′ |
Finally, both versions of the survey asked how the activity could be improved. Responses to this question from all 154 surveys were pooled to give Box 1. The majority (52%) of the responses said that the activity was fine as it is. The remainder suggested various improvements to the activity some of which we will adopt in future lectures (clearer instructions and taking more time to explain the activity).
Improvements (pooled from both surveys; n = 154)
81 (52%) “None,” blank, “fine as it is.”
13 (8%) “Use more flexible wire.”
8 (5%) “Color wire by amino acid properties.”
7 (4%) “Spend more time on the demonstration.”
5 (3%) “Give clearer twisting instructions.”
5 (3%) “Give clearer folding instructions.”
4 (2%) “Allow neighbors to attach wires together.”
3 (2%) “Explain denaturation more.”
The third part of the evaluation measured the longer-term effects of this activity using a follow-up survey conducted at the end of the semester, approximately 7 wk after the activity. This survey was based on survey A, question 1, which asked what students had learned from the activity. To avoid excessive prompting, the follow-up survey questions, shown below, do not mention proteins.
Consider the lecture demonstration from the biochemistry section of the class where you each twisted up a piece of blue wire. As a reminder, the wire is shown below (at greatly reduced scale). (The survey included a reduced version of Figure 1 that only showed the wire; none of the lines or text was included.)
Do you remember this demonstration? (circle one) Yes No
If you answered “Yes” to (1i), please describe what you remember learning from this demonstration. If “No”, go on to (2).
35 Hydrophobic/hydrophilic interactions. Concept (2).
29 Protein folding process. Concept (1).
8 Side chain interactions determine structure.
7 Different-shaped loops represent different types of side chains.
6 Ionic interactions. Concept (2).
3 Vague “proteins.”
3 Blank responses.
2 Backbone versus side chain.
DISCUSSION
The results show that this is a highly successful activity. Students were, in general, able to twist the wire as instructed. Immediately after the activity, students listed the three major concepts as having been learned or represented accurately in the majority of their responses; these concepts were mentioned by only a small minority as being unclear or inaccurately represented. The fraction of students mentioning at least one major concept (75–80%) is very high, given the brevity of the activity. Although self-reported measures are not as reliable indicators of learning as formal content-based pre- and postactivity surveys, our design avoids some of the weaknesses of this type of analysis. Because we used open-ended questions where the students had to name the concepts without prompting, this analysis is less susceptible to novelty, “leading the witness,” and “pleasing the teacher” effects than if we had used multiple-choice content questions or measures of attitude or satisfaction.
The follow-up study shows that students retained several of the learning goals of the activity for at least 7 wk. However, because of the low response rate, the percentage of students remembering the three major concepts should be viewed as an overestimate of retention, because it is likely that students who remembered the activity would be more likely to return the survey form. Even given these reservations, it is clear that this activity had a lasting impact on the students.
In addition to the effects documented above, this activity has other valuable outcomes. It shows students how proteins fold themselves and that this type of self-assembly is possible: although proteins have complex three-dimensional shapes, they are not stamped out of a mold. It provides a concrete illustration of the protein folding process, rather than the endpoint shown by textbook diagrams and molecular visualization. Finally, the demonstration provides a valuable referent for later lectures. As the course progresses, I am able to refer to the wire demonstration as a concrete example when talking about backbone–side chain issues, folding of other proteins, and side chain interactions.
Although there are many positive outcomes from this activity, it should be noted that this highly simplified model of protein folding can lead to important misconceptions, which may need to be specifically addressed with the students. These misconceptions arise from five features of the activity—simplifications inherent in the wire model of protein structure that have important consequences. First, when students twist the side chains along the length of the wire, this implies that the amino acids are formed out of the same material as the backbone rather than being assembled in order from premade monomers. In addition, it implies that hydrophobic side chains are always larger than charged side chains. Second, the activity does not simulate hydrogen bonds; it would be hard to make five distinguishable kinds of loops, and, with only seven amino acids in the chain, it would be difficult to simulate three different types of interactions. Similarly, the activity does not include the effects of side chain shape or covalent bonds between side chains. Third, it suggests that the formation of a hydrophobic core is always the first step in protein folding. Fourth, because the backbone is “inert” in the activity and the polypeptides are short, it is not possible to demonstrate secondary structure. Moreover, these short polypeptides are also incapable of adopting the range of complex three-dimensional shapes formed by full-length proteins. Finally, the wire does not illustrate the amino and carboxy ends of the protein or the directionality of the backbone. Depending on the level of understanding desired by individual instructors, it will be important to discuss some or all of these issues in the context of further treatment of protein folding. This discussion also could lead to a productive conversation about the role of models in science as well as the illustrative and misleading features of any model. Bearing in mind that this activity is designed as an illustration and to provide a referent for later elaboration, rather than as a complete exposition, it is an effective introduction to protein structure and folding.
FOOTNOTES
1 Insulated 18-gauge copper wire is available at most hardware stores. I recommend 18-gauge because thinner gauges are too flexible to hold the final folded shape of the protein, and thicker gauges are too stiff to twist into amino acid side chains. Precut 4-ft lengths of this wire (part no. 9504) are available from Advanced Wire and Cable (Xenia, OH) (www.advancedwire.com) at a cost of $0.30 each (minimum order $50.00). Shorter wires do not allow a sufficient number of amino acids; longer wires are cumbersome and hazardous in a crowded lecture hall.



