An Innovative Plant Genomics and Gene Annotation Program for High School, Community College, and University Faculty
Abstract
Today's biology educators face the challenge of training their students in modern molecular biology techniques including genomics and bioinformatics. The Dolan DNA Learning Center (DNALC) of Cold Spring Harbor Laboratory has developed and disseminated a bench- and computer-based plant genomics curriculum for biology faculty. In 2007, a five-day “Plant Genomics and Gene Annotation” workshop was held at Florida A&M University in Tallahassee, FL, to enhance participants' knowledge and understanding of plant molecular genetics and assist them in developing and honing their laboratory and computer skills. Florida A&M University is a historically black university with over 95% African-American student enrollment. Sixteen participants, including high school (56%) and community college faculty (25%), attended the workshop. Participants carried out in vitro and in silico experiments with maize, Arabidopsis, soybean, and food products to determine the genotype of the samples. Benefits of the workshop included increased awareness of plant biology research for high school and college level students. Participants completed pre- and postworkshop evaluations for the measurement of effectiveness. Participants demonstrated an overall improvement in their postworkshop evaluation scores. This article provides a detailed description of workshop activities, as well as assessment and long-term support for broad classroom implementation.
BACKGROUND AND WORKSHOP DESCRIPTION
Biology faculty face accelerating challenges to introduce their students to modern molecular genetics and bioinformatics techniques. To help address this, with funding from the National Science Foundation (NSF), the Dolan DNA Learning Center (DNALC) designed a professional development project for dissemination of genomics and bioinformatics laboratories to bring faculty up to date with modern plant biology research.
A five-day hands-on workshop on “Plant Genomics and Gene Annotation” was held from June 25–29, 2007, on the Florida A&M University (FAMU) campus. The workshop was advertisednationwide, and 16 faculty from eight states attended the workshop (Figures 1 and 2A). Participants were offered a modest stipend, a free lunch, and refreshments to encourage participation.
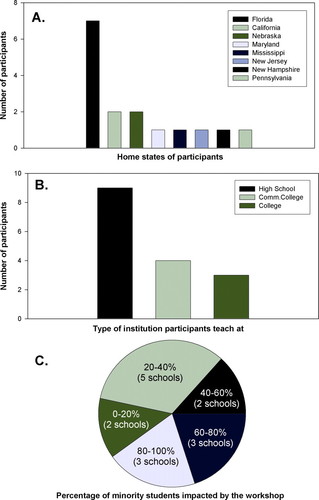
Figure 1. Participant profiles. (A) Home states of participants. (B) Participant institution type. (C) Number of participating schools and their minority student enrollment that was impacted by this workshop.
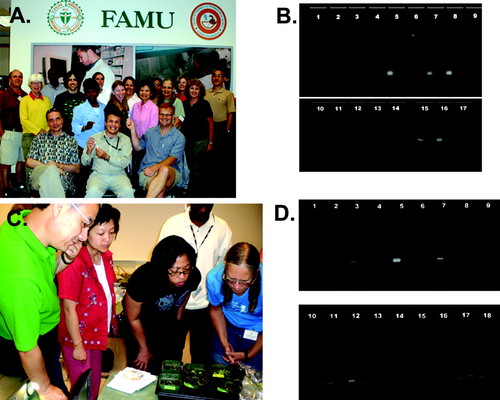
Figure 2. (A) Participants and workshop organizers (seated). (B) Participant-generated DNA gel from Day 1 activities. Note that bands at 271 base pairs or 321 base pairs likely represent BZ allele or mutant BZ allele, respectively. (C) Participants observing Arabidopsis mutants. (D) Participant-generated DNA gel from Day 3 activities. Note that bands at 162 base pairs or 187 base pairs likely represent 35S promoter or tubulin gene, respectively.
The workshop used experiments and bioinformatics activities focusing on plant species, including maize (Zea mays), Arabidopsis (Arabidopsis thaliana), rice (Oriza sativa), and soybean (Glycine max) to illustrate key concepts and techniques (Table 1). These included the relationship of phenotype to genotype, transgenic organisms (as in genetically modified foods), model organisms, gene structure, sequencing, polymerase chain reaction (PCR), genotyping, gel electrophoresis, sequence homology, phylogeny, and gene annotation. The complete list of all equipment, reagents, supplies, and methods is available at the Greenomes website (www.greenomes.org).
| Day 1: Monday, June 25 | ||
| AM | Welcome | Introduction, Workshop Objectives, and Preworkshop evaluation |
| Lab | Zea mays bronze Mutation 1: DNA extraction and PCR | |
| PM | Lab | Zea mays bronze Mutation 2: Gel electrophoresis and analysis |
| Concept | Bioinformatics–What it is and what it can do for you | |
| Computers | Zea mays bronze Mutation 3: Electronic PCR and bioinformatics | |
| Day 2: Tuesday, June 26 | ||
| AM | Concept | Sleuthing for Genes |
| Lab | Arabidopsis clf-2 Mutation 1: DNA extraction and PCR | |
| PM | Lab | Arabidopsis clf-2 Mutation 2: Gel electrophoresis and analysis |
| Computers | Arabidopsis clf-2 Mutation 3: Electronic PCR and bioinformatics | |
| Day 3: Wednesday, June 27 | ||
| AM | Concept | Genetically Modified Foods (GMF) |
| Lab | Detecting GMF 1: DNA extraction and PCR | |
| PM | Lab | Detecting GMF 2: Gel electrophoresis and analysis |
| Computers | Detecting GMF 3: Bioinformatics | |
| Day 4: Thursday, June 28 | ||
| AM | Concept | Gene Structure and Function |
| Computers | Gene Annotation 1 & 2: Meaning, patterns, and finding genes | |
| PM | Seminar | “In Search of Stress Tolerance in Plants at FAMU” by |
| G. Hacisalihoglu, Florida A&M University, Tallahassee, Fla | ||
| Concept: | Evidence for genes | |
| Computers | Gene Annotation 3 & 4: Apollo and building gene models | |
| Day 5: Friday, June 29 | ||
| AM | Concept | Genomes |
| Computers | Gene Annotation 5 & 6: Gene annotation projects and reports | |
| PM | Wrap-up | Postworkshop evaluation, survey, and future directions |
Day 1
The goals of the first day were to: (1) introduce the workshop; (2) introduce the maize transposons through maize bronze mutation experiments (Dooner et al., 1985); and (3) introduce bioinformatics. The bronze allele is caused by insertion of a transposable element first described by Barbara McClintock during her Nobel Prize–winning work on transposons (McClintock, 1951, 1955). This lab served to introduce transposons, genomic complexity, and genomic instability to participants. The workshop began with an interactive plenary session that included welcome remarks by FAMU administrators, introductions by workshop participants and instructors, and workshop objectives (Table 1). Participants were then divided into teams of two and began an experiment to unveil the nature of the mutation in a bronze mutant of maize. The teams performed DNA extractions from wild-type and mutant plants, gene-specific PCR, and gel electrophoresis, thereby determining the genotypic differences between wild-type and bronze mutant Zea mays (Figure 2B). Seeds of maize (Zea mays L.) as well as all reagents were supplied by the DNALC and described at the Greenomes website (www.greenomes.org; Greenomes > Detecting a Transposon Tag in Corn). Maize leaf DNA was isolated as described elsewhere (Edwards et al., 1991; Greenomes website). Genotypes of wild-type (bz) and mutant (bz:Ac) plants as well as primers used are summarized in Table 2. The reaction conditions for the PCR reaction were as follows: 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
| Name/Genotype | Sequence |
|---|---|
| bz-599 (WT) | 5′- CGAATGGCTGTTGCATTTCCAT-3′ (Forward primer) |
| bz-863R (WT) | 5′-ACGGGACGCAGTTGGGCAGGA-3′ (Reverse primer) |
| bz-599 (bz:Ac) | 5′- CGAATGGCTGTTGCATTTCCAT-3′ (Forward primer) |
| Ac-132R (bz:Ac) | 5′-TCTACCGTTTCCGTTTCCGTTT-3′ (Reverse primer) |
| 5′ CLF1 (WT) | 5′-TTAACCCGGACCCGCATTTGTTTCGG-3′ (Forward primer) |
| 3′ CLF2 (WT) | 5′-AGAGAAGCTCAAACAAGCCATCGA-3′ (Reverse primer) |
| 5′ CLF1 (Mutant) | 5′-TTAACCCGGACCCGCATTTGTTTCGG-3′ (Forward primer) |
| 3′ Ds (Mutant) | 5′-GTCGGCGTGCGGCTGGCGGCG-3′ (Reverse primer) |
| 35S-5′ (Promoter) | 5′-CCGACAGTGGTCCCAAAGATGGAC-3′ (Forward primer) |
| 35S-3′ (Promoter) | 5′-ATATAGAGGAAGGGTCTTGCGAAGG-3′ (Reverse primer) |
| Tub5 (Tubulin) | 5′-GGGATCCACTTCATGCTTTCGTCC-3′ (Forward primer) |
| Tub3 (Tubulin) | 5′-GGGAACCACATC ACCACGGTACAT-3′ (Reverse primer) |
| g4026–5′ (Marker) | 5′-GGGGTCAGTTACATTACTAGC-3′ (Forward primer) |
| g4026–3′ (Marker) | 5′-GTACGGTTCTTCTTCCCTTA-3′ (Reverse primer) |
| H77224–5′ (Marker) | 5′-GGATTTGGGGAAGAGGAAGTAA-3′ (Forward primer) |
| H77224–3′ (Marker) | 5′- TCCTTAGCCTTGCTTTGATAGT-3′ (Reverse primer) |
Bioinformatics uses computational-based tools to solve biology-related problems. The workshop participants were introduced to bioinformatics algorithms used for gene or promoter finding, sequence alignment, and gene annotation. Bioinformatics exercises complemented the bench work and helped participants to develop an understanding of the cause of the bronze mutant phenotype. The bioinformatics exercises included mapping a Ds insertion and identifying the inactivated protein (Greenomes > Detecting a Transposon Tag in Corn). More specifically, participants: (1) used BLAST to find DNA sequences matching the primers in biological databases (National Center for Biotechnology Information, NCBI; www.ncbi.nlm.nih.gov); (2) identified the Bz amino acid sequence; and (3) determined the chromosomal location and function of Bz and Ds using NCBI's GenBank datasheets and Map Viewer tool.
Day 2
During the second-day session, “Sleuthing for Genes,” participants: (1) investigated genotypic and phenotypic relationships while analyzing a mutation in Arabidopsis curly leaf (clf, Kim et al., 1998) obtained by transposon-induced mutagenesis; and (2) identified sequences in biological databases for the CLF gene and protein. This lab illustrates how deliberate transposon-induced mutagenesis screens allow fast identification of genes required for biological processes (Figure 2C). Seeds of Arabidopsis as well as all reagents were supplied by the DNALC and described at the Greenomes website. DNA was isolated as described elsewhere (Edwards et al., 1991; Greenomes website). The reaction conditions for the PCR reaction were as follows: 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s. Participants completed DNA extraction and analysis of clf-2 and wild-type plants, mirrored with identification and analysis of published data about clf-2 using online databases and bioinformatics tools.
Bioinformatics exercises complemented the lab with insights into the nature and structure of the mutated gene and its protein product. Furthermore, participants discovered gene features to be used in gene mining from genomes of other organisms. More specifically, participants: (1) used BLAST to find DNA sequences related to CLF1/CLF2 in databases; (2) identified the CLF amino acid sequences and CLF1/CLF2 amplicon; (3) used Map Viewer to determine the chromosome location of the CLF gene; (4) determined the insertion site of the Ds transposon; and (5) used BLAST to determine the function of CLF protein (Greenomes > Detecting a Transposon Tag in Arabidopsis). All bioinformatics exercises including videos and animations can be reached at a complementary Bioinformatics website (http://bioinformatics.dnalc.org/clf).
Day 3
During the third day, on “Genetically Modified Organisms,” participants: (1) isolated DNA from various plants, including soybeans, and dry prepared foods (such as cereals and crackers); (2) used a PCR assay to identify which samples included transgenic DNA; and (3) practiced their newly developed bioinformatics skills to discover the functions of transgenes in their plant and food products. Seeds of wild-type and Roundup Ready soybean (Padgette et al., 1995) were supplied by DNALC. Participants supplied dried food products and were encouraged to bring those containing soybean, as soybean is one of the most widely grown genetically modified (GM) crops (Stalker et al., 1988). All reagents were supplied by the DNALC and described at the Greenomes website (Greenomes > Detecting Genetically Modified Foods). DNA was isolated as described elsewhere (Edwards et al., 1991; Greenomes website). The reaction conditions for the PCR reaction were as follows: 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s (Vollenhofer et al., 1999). A representative gel generated by participants on Day 3 is shown in Figure 2D.
The participants explored which genes have been introduced into plants using bioinformatics. Bioinformatics exercises and discussion showed participants the process of genetic modification, introducing transformation and selection methods, gene(s) of interest and their applications in the target organisms, and promoters allowing expression in plants. More specifically, participants used BLAST (NCBI) to identify the 35S promoter in transgenes and then investigated which genes have been engineered for expression under the control of the promoter (Greenomes > Detecting Genetically Modified Foods by PCR). Methods for producing transgenic plants and their regulations were discussed.
Day 4
The fourth day sessions on “Gene Structure and Function” included: (1) introducing genome annotation in nucleotide and protein levels, and (2) analyzing and interpreting data from Days 1–3. Participants investigated patterns in DNA and used computer programs to predict genes in DNA sequences. Participants were then introduced to gene annotation and explored predicted gene models and biological evidence for genes, such as cDNA sequences using Apollo, a gene annotation program (Lewis et al., 2002). Bioinformatics exercises included predicting the size of CAPS (cleaved amplified polymorphic sequences) amplicons with BLAST, and mapping AGO1 (argonaute; Bohmert et al., 1998) and CAPS markers electronically to identify homologous regions of chromosomes (Greenomes > Linkage Mapping a Mutation in Arabidopsis).
Another component of Day 4 activities was a research seminar entitled “In Search of Stress Tolerance in Plants at FAMU” (Hacisalihoglu et al., 2007; Hacisalihoglu and Kochian, 2003), which served to introduce current application of genetics and plant molecular biology to economically important agricultural issues. More specifically, the research seminar helped participants to discuss the practical applications of modern plant biology in areas such as plant nutritional quality and increased plant resistance against bacterial diseases.
Day 5
The last session, on “Genomes”: (1) continued analyzing gene annotation, and (2) included postworkshop evaluations. Participants completed independent gene annotation projects using Apollo, annotating predicted genes of their choice from the Rice genome. More specifically, Day 5 involved making gene models, synteny (conservation of adjacent genes), improvement of gene models by using ESTs, inserting a transcript, as well as alternative splicing. The Dynamic Gene website (www.dynamicgene.org; Dynamicgene > Annotation) includes online protocols, detailed tutorials, and multimedia resources supporting these activities.
PARTICIPANTS, LOCATION, AND MATERIALS
Participants
A total of 16 highly qualified high school, community college, and university faculty were selected for the workshop from 34 applicants (Figures 1A, 1B, and 2A). Selection criteria included current teaching assignments, participation in curriculum development, experience with laboratory teaching, minority status, as well as percentage of minority students in participants' schools. An additional 10 applicants were placed on a waiting list. The profile of the workshop participants is summarized in Figure 1. Sixty-eight percent of the participants had master's degrees, whereas 13% had doctorates. The remaining participants had a B.S., B.A., (13%), or M.B.A. (6%). About 38% of participants were from minority groups. All participants had experience in molecular biology laboratory instruction as well as using the Internet for classroom teaching (data obtained from preworkshop applications). Fifty percent of the participating schools had over 40% minority student enrollment (Figure 1C).
Location
The workshop was conducted on the campus of FAMU in Tallahassee, FL. FAMU is a historically black university with more than 95% African-American student enrollment. Holding the workshop at FAMU greatly increased the visibility of modern plant biology research among underrepresented minorities, as the workshop was featured on the FAMU main website (www.famu.edu) and DNALC website (www.dnalc.org). Underrepresented minorities constituted approximately 38% of the workshop participants.
Materials
The majority of the equipment and all supplies for the workshop were provided by the DNALC, including thermal cyclers, pipettors, a microcentrifuge, precast agarose gels, and enzymes. Access to additional equipment was provided by FAMU including refrigerators, freezers, water baths, and a microwave oven. For bioinformatics labs, participants either used DNALC or personal laptop computers and software. All specialized software used in the workshop is available free on the Internet (Greenomes and Dynamic Gene websites > Resources), increasing accessibility of the curriculum.
MEASURING SUCCESS AND RESULTS
Participants were given pre- and postworkshop evaluations to assess the effectiveness of the workshop. Sample questions and answers are presented in Table 3. A closed-book multiple-choice questions (MCQ) test was used for assessment. The evaluation was composed of 20 MCQs with four multiple-choice answers of which one is correct. MCQs were machine-graded with an optical scanner at FAMU. The data were subjected to analysis of variance and separated using Schaffe procedure at P < 0.05 (SPSS, Chicago, IL).
| 1. What is the haploid chromosome number of model plant Arabidopsis thaliana? | |||
| a) 6 chromosomes | b) 22 chromosomes | c) 10 chromosomes | d) 5 chromosomes |
| 2. ″Ac″ and ″Ds″ transposons in maize are: | |||
| a) Mutations | b) Promoters | c) Transposons | d) 5 transcripts |
| 3. ″Genetic linkage″ in plants is: | |||
| a) Mitochondrial genes | b) Polygenes | c) Genes on the same chromosome | |
| 4. What does ″Bt corn″ stand for? | |||
| a) A GMO corn | b) A high yield corn | c) A sweet corn | d) A European corn |
| 5. What does ″RR soybean″ stand for? | |||
| a) A dominant gene | b) Homozygous allele | c) Parents of soybean | d) Roundup ready |
| 6. The acronym ″GMO″ stands for______________________? | |||
| a) A type of organic food | b) Genetically modified organisms | c) Parental plants | |
| 7. Name a computer program used for ″gene annotation″ in plants. | |||
| a) Firefox | b) Apollo | c) Challenger | d) Dreamweaver |
| 8. The most important finding of Barbara McClintock was: | |||
| a) QTLs | b) Arabidopsis mutations | c) pea mutations | d) transposable elements |
As seen in Table 4, there was an overall improvement in scores for the postworkshop evaluation compared with preworkshop evaluation. The postworkshop evaluation scores increased more than 35 percentage points (pp) for community college faculty, 27 pp for university faculty, and 33 pp for high school faculty when compared with preworkshop evaluation scores. Furthermore, minority participants had 34-pp increases in their postworkshop scores compared with all others (Table 4). However, there was no significant difference between postworkshop scores of minority and nonminority participants.
| Participant groups | Preworkshop evaluation (%) | Postworkshop evaluation (%) point difference | Percentage | Number of participants |
|---|---|---|---|---|
| High school | 62a | 85ab | 23c | 8 |
| Community college | 58a | 93a | 35a | 4 |
| Four-year college | 43a | 70b | 27b | 4 |
| Significance | NS | * | * | |
| All participantsa | 54b | 82a | ||
| Minority | 46b | 80a | 34a | |
| Nonminority | 60a | 80a | 20b | |
| Significance | * | NS | * |
The success of the workshop is best conveyed by the written comments of some participants at the end of the workshop:
“Well-organized, appropriately paced, logical progression of skills and topics. Sensitivity to participant background, exciting environment, and hospitality.”
“The resources (CDs and websites) were wonderful, and I look forward to using them as teaching tools for my molecular genetics and biology courses.”
“The workshop was a great match for my needs. I enjoyed all aspects of the course: presentations, speakers, labs.”
“We thank the organizers for taking such good care of all the details. I have got some good hands-on experience with searching for mutations in various plants. I will have so much to tell my students about the opportunities that await them in biology.”
“This has been a tremendously helpful workshop. I very much appreciate the opportunity to participate and everyone who made it possible.”
“I appreciate everything that organizers and NSF did to make this workshop possible. I look forward to taking my new knowledge back to my students and coworkers.”
CONCLUSIONS
This article describes and summarizes outcomes of a “Plant Genomics and Gene Annotation” faculty workshop. This workshop gave participants a deeper understanding of modern methods in genomics and bioinformatics. Participant surveys as well as the evaluation regimen indicate that the design and format of the workshop was effective at facilitating active learning using hands-on in silico and in vitro experiments. One of the strengths of the workshop was the ability to reach underrepresented minority faculty. Pre- and postworkshop evaluation scores indicated that participants expanded their knowledge of plant molecular genetics. A significant difference was found between community college faculty and four-year college faculty. It is reasonable to assume that the workshop was designed for high school and community college faculty rather than four-year college faculty. Furthermore, there was no significant difference between postworkshop scores of minority and nonminority participants. This result shows that the workshop was very effective to compress all differences that existed at the beginning of the workshop (Table 4).
A 1.5-d follow-up workshop will be conducted at FAMU during the spring of 2008, allowing participants to refresh their newfound knowledge and address concerns after implementing the current workshop material in their teaching. FAMU will then receive an equipment package including a thermocycler, centrifuge, UV-lightbox, and pipettors to establish an equipment loan program. This will provide participating faculty everything needed to conduct molecular biology and genomics experiments with their students. Although we are encouraged by the success of the Plant Genome workshop, a couple of limitations need to be acknowledged. First, we recognize that positive outcomes of this workshop are dependent on use of workshop activities by participants in their teaching. Second, because of the popularity of the workshop, we were unable to accommodate the demand. We would therefore recommend more Plant Genomics workshops, both at FAMU and other locations nationwide.
ACKNOWLEDGMENTS
We acknowledge the NSF for funding the workshop, and workshop participants for their hard work and contribution. G. H. thanks M. Timmermans (Cold Spring Harbor Laboratory) and M. Scanlon (Cornell University) for the faculty fellowship opportunity and R. Reams (FAMU) for providing access to additional equipment and assistance with the workshop set-up. A fellowship for G. Hacisalihoglu, the workshop at Florida A&M University, and development of the Dynamic Genes Internet site were supported by grants from the National Plant Genome Initiative of the NSF. The Greenomes curriculum and Internet site were developed with support from NSF's Course, Curriculum, and Laboratory Development Program.



