Mathematical Biology Modules Based on Modern Molecular Biology and Modern Discrete Mathematics
Abstract
We describe an ongoing collaborative curriculum materials development project between Sweet Briar College and Western Michigan University, with support from the National Science Foundation. We present a collection of modules under development that can be used in existing mathematics and biology courses, and we address a critical national need to introduce students to mathematical methods beyond the interface of biology with calculus. Based on ongoing research, and designed to use the project-based-learning approach, the modules highlight applications of modern discrete mathematics and algebraic statistics to pressing problems in molecular biology. For the majority of projects, calculus is not a required prerequisite and, due to the modest amount of mathematical background needed for some of the modules, the materials can be used for an early introduction to mathematical modeling. At the same time, most modules are connected with topics in linear and abstract algebra, algebraic geometry, and probability, and they can be used as meaningful applied introductions into the relevant advanced-level mathematics courses. Open-source software is used to facilitate the relevant computations. As a detailed example, we outline a module that focuses on Boolean models of the lac operon network.
INTRODUCTION
In the last decade, the field of life sciences has undergone revolutionary changes spanning remarkable discoveries at all levels of biological organization—molecules, cells, tissues, organs, organisms, populations, and communities. A salient trait of these advances is the increased need for statistical, computational, and mathematical modeling methods. Scientific instruments are now, by orders of magnitude, more sensitive, more specific, and more powerful. The amounts of data collected and processed by these new-generation instruments have increased dramatically, rendering insufficient the traditional methods of statistical data analysis. Nowhere, however, have the problems of amassing huge amounts of data been more clearly demonstrated than in attempts to unravel the secrets of genetic mechanisms.
For example, automated DNA sequencing has given rise to an information explosion, and the challenge now is to extract meaning from all of this sequence information. The quest to better understand temporal and spatial trends in gene expression has led us to search for DNA sequences that have been conserved over time in a large number of species. The existence of such conserved strings in different species suggests that these sequences may perform fundamental functions in the genome and thus be critical to our understanding of life on earth. However, determining candidates for DNA sequences that have been conserved over time across different species is a tremendous task, because the human genome alone is approximately 3 billion base pairs. Comparing across species then requires comparisons of further billions of sequences, over thousands of species. The sheer size of the data sets suggests that appropriate use of mathematical models coupled with statistical methods for data analysis and inference will play an irreplaceable role in contemporary biology. Frequent announcements of the sequencing of additional organisms, such as the rhesus macaque (Gibbs and the Rhesus Macaque Genome Sequencing and Analysis Consortium, 2007) and the domestic horse (Wade et al., 2009) demonstrate that the complexity of the data sets is continually growing; thus, future advances in molecular biology will need to rely even more heavily on the use of mathematical methods.
Similarly, the field of molecular systems biology has emerged as equally mathematically driven (Robeva, 2010). Broadly defined, this is a field that examines how “… large numbers of functionally diverse, and frequently multifunctional, sets of elements interact selectively and nonlinearly to produce coherent behavior” (Kitano, 2002). Thus, organismal function and behavior is determined by a tremendously complex set of interactions (e.g., protein–protein, protein–DNA, protein–RNA), and the complexity of the interactions requires the assistance of mathematics if we are to understand how living things function. Understanding these interactions will enable greater progress against conditions such as heart disease, cancer, and diabetes. A recent proposal for a new national initiative (toward “the New Biology”) identifies health issues as one of four key areas where a systems biology approach and improvements in mathematical and statistical modeling will be prerequisites for progress: “Although there are increasing efforts to apply quantitative approaches to biological questions, more must be done to transform biology from its origins as a descriptive science to a predictive science. We will ultimately be limited in our ability to deploy biological systems to solve large-scale problems unless we significantly deepen our fundamental understanding of the organizational principles of complex biological systems, a staggeringly difficult challenge. The growth of the New Biology will be dramatically accelerated by developing frameworks for systematically analyzing, predicting, and modulating the behavior of complex biological systems.” (A New Biology for the 21st Century; National Research Council [NRC], 2009). The challenge is to combine the rich but disparate insights of molecular biology into a conceptual framework that better allows us to see the overall structure of molecular (and other) mechanisms. Mathematical models have proved to be indispensable in this regard. Indeed, the assessments that “… the main push in biology during the coming decades will be toward an increasingly quantitative understanding of biological functions …” (Mathematics and 21st Century Biology; NRC, 2005) and that “… the traditional segregation in higher education of biology from mathematics and physics presents challenges and requires an integration of these subjects …” (Rising Above the Gathering Storm; National Academy of Sciences, National Academy of Engineering, and Institute of Medicine, 2007) are now widely accepted and a range of diverse mathematical methods are now routinely used to seek answers to questions from systems biology.
Preparing students of our educational systems to meet these challenges is consequently a crucial national need. However, although the interaction between mathematics and contemporary biology increasingly requires the use of diverse mathematical methods, responses through curricular changes at the undergraduate level have been largely constrained to the application of difference and differential equations to model the dynamics of biological systems, possibly attributable to the following factors: 1) these types of models have been extremely successful historically in providing answers to questions in ecology, epidemiology, physiology, and pharmacology; 2) once formulated, the existence of rich mathematical theory in the field of ordinary differential equations allows for standard analyses of such models; 3) as calculus and differential equations courses most often form the core of the undergraduate mathematics major, the study of such models provides natural extensions of the course curricula; and 4) a relative lack of historical interactions between biology and mathematics (compared with prominent connections between physics, engineering, and chemistry with mathematics) means there are relatively few mathematicians who are themselves trained in biology, and of those who are, most will have encountered biology through calculus (i.e., mathematical analysis)-based programs.
Thus, of the two broad groups of mathematical methods currently used to organize insights into contemporary molecular biology—analytical and algebraic—undergraduate mathematical biology curricula are biased primarily toward the analytical, calculus-based1 approaches. Methods in the algebraic group include elementary discrete mathematics, graph theory, probability, linear algebra, abstract algebra, and polynomial algebra and algebraic geometry. In this discussion, we refer to this selection of (noncalculus-based) methods as modern discrete mathematics. Particularly notable in this group is the field of algebraic geometry. At its most elementary level, this is a classical subject that captures mathematical patterns and relationships through geometric figures (varieties) that can be described as sets of common solutions to polynomial equations.2 To emphasize the parallel with linear algebra, which studies solution sets to systems of linear equations, the term polynomial algebra is also used. Combining methods from algebraic geometry/polynomial algebra with concepts from probability and statistics is the province of algebraic statistics.
Aspects of modern discrete mathematics and algebraic statistics have recently made a significant impact on molecular biology, much as calculus-empowered population biology and epidemiology have had in the early 20th century. Examples include finite dynamical systems models of the metabolic network in
We believe the notable absence of modern discrete mathematics from the undergraduate mathematical biology curricula can be attributed primarily to a lack of appropriate materials rather than inaccessibility of the essential underlying mathematical concepts. Indeed, elementary topics in modern discrete mathematics and the underpinnings of algebraic statistics (including topics from discrete mathematics, matrix theory, graph theory, linear and polynomial equations, and basic probability and statistics) constitute the bulk of general education mathematics courses, whereas more advanced topics (i.e., in linear algebra, graph theory, and modern algebra) permeate the noncalculus-based math major curriculum. As indicated by the popularity of texts such as “Ideals, Varieties, and Algorithms” (Cox et al., 2007), now in its seventh printing, entire courses in algebraic geometry have become popular at many institutions. One of this paper's authors (T. H.), whose mathematical research utilizes aspects of algebraic geometry, has run well-received courses and seminars in algebraic geometry and related topics to mixed audiences of preservice high school teachers, undergraduate mathematics majors, graduate students, and mathematics faculty. Reinhard Laubenbacher's Discrete Mathematics Group at the Virginia Bioinformatics Institute (VBI) has run two biomathematics workshops for in-service high school teachers connecting topics in modern discrete algebra to the Virginia public schools' required Standards of Learning (Martins et al., 2008).
For the collection of modules outlined in this article, our goal is to integrate appropriate concepts from modern discrete mathematics and algebraic statistics with the relevant molecular and systems biology for use in the standard mathematics and biology curricula.
PEDAGOGICAL FRAMEWORK
The collection of modules is designed around the principle of project-based learning (PBL), focusing on problem solving that emphasizes important mathematical concepts and methods in the context of essential questions raised in modern biology. This pedagogy has been proved to increase students' depth of understanding of the material as well as students' ability to make use of their knowledge in new situations (Boaler, 1998). Furthermore, in selecting the project topics, we strived to follow a fundamental principle of the PBL approach: selecting authentic, real-world problems that are (or have been) important for advancing biology while providing a feasible venue for student learning in both mathematics and biology. The goal is to enable students to think in terms of mathematical models and motivate them to develop and further apply their mathematical and modeling skills.
In their past work, some of the authors have used the PBL approach successfully to develop a separate, primarily calculus-based collection of projects that have been published as a “Laboratory Manual of Biomathematics” (Robeva et al., 2007b). This manual can be used together with the textbook “Invitation to Biomathematics” (Robeva et al., 2007a) or as an independent project book. For this current collection, we adopt the same approach, which is summarized next. A detailed description has been published (Robeva, 2009).
The modules for the current project are directed toward the interface of mathematics and biology and are intended for use in both mathematics and biology courses. As already mentioned, many significant research projects now require real cross-disciplinary collaboration and our aim is to develop project modules that exemplify these experiences in the classroom to the extent possible. This includes the use of specialized software to facilitate some of the computationally heavy techniques for which hand computations are not feasible. The modules aim to:
Improve mathematics students' understanding of the role of mathematical models in the life sciences on the one hand, and biology students' mathematical skills on the other. This includes the ability to look at an unfamiliar problem arising from biology, understand the need for using mathematical methods to address the problem, and recognize how mathematical approaches (spanning a wide range of mathematical techniques and, at the introductory level, fully accessible to biology students who have not yet taken calculus) can be instrumental in the search for answers.
Reinforce students' mathematical background by exposing them to current ideas and by presenting the mathematical topics they have encountered before, but from novel points of view. In a number of traditional programs, students may be exposed to some mathematical concepts in such a limited or abstract fashion that they are aware of encountering the material again only in subsequent mathematics courses, if at all. This may, at times, convey the false impression that mathematics is present in the undergraduate curriculum and requirements solely as an abstract logical and algebraic exercise. The project modules emphasize creative applications.
Introduce new mathematical tools in the context of engaging problems. In our past work, we found that PBL use not only improved students' abilities to use their knowledge in new situations but also better equipped them to learn and understand new mathematical content in the process of engaging with the project (Robeva, 2009).
The foci and arrangement of the modules allows for their use in isolated class meetings or in units of 2 to 3 weeks, thus maximizing the utility of the modules for the faculty choosing to use them. Biology drives the presentation with the appropriate mathematical theory presented in the context of the problem-solving process. The low-level mathematics that is initially necessary to address aspects of the problem is introduced in detail. Thus, the early parts of the module, paired with the description of the general mathematical background and hands-on exercises, are generally appropriate for use in existing biology courses including genetics, evolution, and cell and molecular biology. The subsequent progression of questions in the modules then leads to more challenging mathematical questions, thus making them appropriate for use in the upper-level mathematics courses such as linear algebra and abstract algebra. The modules are generally open-ended, listing questions for student exploration similar to those examined in the project together with relevant references for further reading.
The mathematics is introduced and explained in detail and is not treated as a black box. To complete the module (or those parts of it chosen by the instructor), students need to work through both the biology and the mathematics. Biology students will need to understand the mathematics and mathematics students will need to understand the biology. But as the mathematics becomes progressively more challenging, the use of the modules in conventional biology courses will be focused on the early parts of the modules. It will be unrealistic to expect biology students to understand the details of many of the high-level, abstract mathematical concepts to which the projects lead, but we are aiming at framing questions so that biologists can see the issues involved and appreciate the specific benefits of using mathematical approaches to answer the questions. This approach aims to respond to one of the basic rules of interdisciplinary collaboration: not everyone on the interdisciplinary team needs to know everything, but team members should know enough from the other disciplines to be able to effectively communicate with one another.
MODULE DESIGN AND METHODS
Our modules pull from numerous advances made in systems biology, genomics (investigating the function and structure of genes and genomes), and phylogenetics (identifying and understanding evolutionary relationships among the various kinds of life on earth), facilitated by the use of mathematical and computational methods. The following essential biological questions are used to provide the main strands intertwined to form the modules: 1) Given partial information about the functional structure of a biological network, what types of models are appropriate to more fully capture the network properties and function? 2) Given data reflecting the time-evolution of a biological network, what internal mechanisms are responsible for the observed behavior? and 3) Given a collection of DNA sequences, what underlying forces are responsible for the observed patterns of variability?
Question 1 is the most common question in mathematical modeling, with the aim to develop models designed to understand system properties based on previous knowledge of structure. Such models are typically parsimonious, including a minimal number of key functional elements and interactions that describe the structure or the principal dynamics of the system. Two types of mathematical models have been used successfully to organize insights of molecular biology and capture network structure and dynamics: 1) discrete- and continuous-time models built from difference equations or differential equations, which focus on the interaction kinetics; and 2) discrete-time algebraic models built from functions of variables with values from a finite set S, which focus on the logic of the network variables' interconnections.
The special case of S = {0, 1} corresponds to the Boolean networks model proposed by Kauffman (1969), in which model variables are discretized to values from the set S and considered to be either present or absent. The state space of such systems can be represented by directed graphs (digraphs) with 2n vertices, where n is the number of variables (each state corresponds to an n-tuple of 0's and 1's). In the Example section below, we describe this concept in more detail.
Boolean networks represent a special case of a more general type of dynamical systems referred to as polynomial dynamical systems (PDS), wherein the dependency diagram between the network variables is defined by functions that are polynomials of these variables. Any finite dynamical system can be represented as a PDS (see Comparing Algebraic and Calculus-Based Models of the Lac Operon), so examining the properties of such systems is of particular importance.
Question 2 refers to reverse engineering where information about the structure of a biochemical network is derived from time course data without prior knowledge of its topology. There is growing evidence (e.g., see Davidson, 2002; Wang and Cherry, 2002; Laubenbacher and Stigler, 2004; Laubenbacher and Mendes, 2006; Dimitrova et al., 2007) that time-discrete dynamical systems over a finite state space S (commonly referred to as finite dynamical systems) may be better suited to capture key features in certain types of gene regulatory networks. In fact, it has been shown that methods from modern discrete mathematics can be used successfully for reverse engineering (Dimitrova et al., 2007).
Question 3 requires the use of methods from genomics and phylogenetics. For example, in complex organisms, genes do not occur as unbroken DNA sequences but are split into pieces, called exons, with intervening sequences of noncoding DNA, called introns. Only the exons carry the genetic code, yet ∼96% of a genome may consist of introns (Watson, 2003, pp. 109–110). The problem of genome annotation is to 1) parse genomes into DNA sequences that have some identifiable characteristic and then 2) to attach biological information to those identified sequences; that is, having identified a DNA sequence as a gene, one wants to determine what biological or biochemical functions the gene regulates in the organism.
Traditional statistical and computational methods have always been essential to the problems of gene annotation, but new approaches based on algebraic statistics and modern discrete mathematics are proving to be equally important. For example, hidden Markov models (HMMs) are probabilistic models capable of capturing both known features (such as genes) and hidden features of the data under analysis (such as the introns and exons of genes). HMMs are central tools for modern genome data analysis and are used routinely for genome annotation by data sequencing centers (Pachter and Sturmfels, 2007). HMMs can be presented concisely and conceptually via graphs and figure prominently in biological applications of algebraic statistics, because HMMs can be reinterpreted geometrically as varieties. The enormous advances in computational algebraic geometry over the past three decades can thus be applied to analyze the genome annotation problem (Pachter and Sturmfels, 2005; Pachter and Sturmfels, 2007). Related questions of equal importance where HMMs also play a key role are those of sequence alignment where the problem is to determine the fewest number of allowable changes (edits) that will account for the mutational pathway from one genetic sequence to another. When comparing across different species, the problem of multiple sequence alignment can be represented and explored through phylogenetic tree graphs in which evolutionary links between these species are inferred from conservation of DNA sequences across species.
Expanding access to the set of mathematical methods used in modern molecular biology so as to incorporate modern algebra, geometry, and other discrete structures, coupled with optimization theory and increasingly sophisticated probabilistic and statistical modeling techniques, follows a parallel trend in modern mathematics. This trend has expressed itself through reforms at the K–12 level such as those appearing in the National Council of Teachers of Mathematics standards (http://standards.nctm.org), at the undergraduate level through the success of very accessible undergraduate texts such as Ideals, Varieties, and Algorithms (Cox et al., 2007) that require little prior mathematical background, and at the research level through the ascension of accompanying theories such as coding theory, advanced linear algebra, representation theory, Lie theory, graph theory, and topology empowered through their ties with, and applications to, modern computing. The strength of these theories lies (like the best of biological models) in their organizational power, and in their ability to capture patterns of increasing complexity and mathematical and physical sophistication, even as they stand firmly rooted in elementary principles such as functions, elementary geometry, algebraic operations, linear equations and matrices, the graphing and long division of polynomials of a single variable, and number systems.
MODULE TOPICS
We now present a brief outline for the educational modules that are currently under development. This list of projects may expand in the future to accommodate further projects under consideration.
1. Discrete Mathematical Models of Gene Regulation and of the Lac Operon
The module focuses on developing algebraic models of gene regulation. It introduces the concepts of finite dynamical systems, polynomial systems, and dynamical systems over finite fields and applies them to creating a model of the lac operon. In addition, models of the lac operon with delay are considered. The ability of the lac operon to exhibit bistability also is examined. For relatively simple systems involving only a few variables (such as the Boolean system in the example below), the complete transition diagram can be computed and depicted as a directed graph. In such cases, the graph also shows whether the system has fixed (equilibrium) points or limit cycles or if it is bistable. For systems composed of larger numbers of variables, visual verification is not feasible as the size of the state space increases exponentially with the number of variables. For such systems, the search for answers leads to more advanced mathematical concepts including systems of polynomial equations, algebraic varieties, and Groebner bases.
2. Comparing Algebraic and Calculus-based Models of the Lac Operon
A significant number of discrete and differential models of the lac operon are now available in the literature. In this module, we examine the similarities and the differences between some of these models, including a minimal differential equations model of the lac operon (Santillán and Mackey, 2008) and a PDS counterpart based on the same wiring diagram. In case module 1 above is used with students who have already taken calculus (in this case, we believe two semesters of calculus is appropriate), this module could be used as a follow-up. Particular emphasis is placed on the fact that differential equations models are quantitative and that PDS models are qualitative in nature, discussing the advantages and disadvantages of each of these approaches. We revisit the question of bistability and examine differential equations models and PDS models, including models with delay, that exhibit the bistability property. Some of these approaches are quite general and can be applied to other systems, including the lambda phage switch (Hinkelmann and Laubenbacher, 2009).
3. Reverse Engineering of Biochemical Networks
This module examines methods that allow information about the structure of a biochemical network to be derived from time course data without prior knowledge of the network structure (topology). We show specifically how discrete dynamical systems over a finite state space S are well suited to capture key features in certain types of gene regulatory networks. The special case of S = {0, 1} corresponds to the classical Boolean networks models proposed by Kauffman (1969), in which gene expression is discretized to values from the set S, hence considered to be either present or absent. The mathematical level of sophistication spans a wide range of topics from elementary logic and graph theory to advanced abstract algebra and computational algorithms. Some of the examples are related to models of the lac operon network included in module 1.
4. When Does Evolution Occur? (and How Mathematics Helped Answer This Question)
This module begins with a discussion of the classical work of Luria and Delbrück on mutation of bacteria from virus sensitivity to virus resistance. We next examine the fluctuation test that grew out of this work and provided the first experimental proof (by using probabilistic models and statistical approaches) that bacterial mutations follow a Darwinian and not a Lamarckian model. Interestingly, Max Delbrück also was the first one to draw attention to the fact that biological systems can exhibit bistability (Delbrück, 1949). Novick and Weiner (1957) not only experimentally demonstrated this feature for the lac regulatory network but also showed that the state of a single cell (induced or uninduced) could be transmitted through several generations. This provided one of the simplest examples of phenotypic, or epigenetic, inheritance. We next discuss the role of epigenetic inheritance in relation to the Darwinian model inferred by Luria–Delbrück, and the regulation of protein synthesis. This same topic is also related to modules 7 and 8 below, exploring the link between epigenetic states of CpG islands and cancer.
5. Linear Algebraic Approaches to Metabolite Conservation
Metabolic pathways, such as glycolysis, form the biochemical drivers for life for systems at the cellular and organism level. Conservation relationships for metabolic concentrations are linear dependencies that can be analyzed and modeled by special connectivity matrices (i.e., stoichiometric matrices) in a manner that resembles classical problems in electrical circuit analysis. Basic concepts from linear modeling and introductory linear algebra (e.g., Gaussian elimination, linear combinations and independence, fundamental subspaces), as well as more advanced topics (e.g., inner product spaces and singular value decomposition) are brought to bear to examine these molecular biochemical networks, and topics from elementary numerical linear algebra are invoked to understand fundamental constraints on using these methods at the genomic level. Such mathematical models extend current biological intuition and suggest mechanisms for understanding how living systems maintain steady states and fight or fall to disease, as well as the proper design of medical interventions.
6. Geometry of Phylogenetic Tree Reconstruction
This module explores topics in elementary graph theory, linear algebra, modern and polynomial algebra, combinatorics, linear programming, and algebraic statistics through applications to a central and profound problem in biology: the determination of hereditary relationships among organisms through the alignment of DNA sequences from creatures currently in existence. An added new twist nowadays is genetic data extracted from organisms that have long been extinct, including the use of protein analysis from fossilized Tyrannosaurus rex bones to show an evolutionary link to chickens (Organ et al., 2008). Visually appealing diagrams that can take on a number of forms, phylogenetic trees are a commonly used model for representing evolutionary relationships among taxa (e.g., species, organisms, or genes) as the tips or leaves of a tree, with branches joining two or more taxa at an internal node (branch point) to indicate that they share a common ancestor. Mathematically, phylogenetic trees can be viewed as graphs and also identified as points in an appropriate geometric context. This module uses both perspectives to consider a number of mathematical approaches, including neighbor-joining, balanced minimum evolution, and singular value decomposition methods, to the problem of recreating the best phylogenetic tree from only the partial data associated to the leaves given by DNA sequence alignments.
7. Codon Usage, CpG Content, and Genome Signature
Fundamental concepts from probability and statistics form the mathematical core of this module on DNA and the genetic code, codon usage, codon usage bias, and related concepts. We examine an accessible number of basics from genomics and molecular biology leading to questions of great historical import (e.g., debates over the applicability of Darwinian evolution at a molecular level), as well as to current research frontiers. Modeling with polynomial algebra or algebraic geometry by way of algebraic statistics also may be used to address some of the questions raised in this context. This module also is related to module 8 below for determining computational methods for locating CpG islands on the genome.
8. HMMs in DNA Sequence Analysis
HMMs have been used successfully since the 1980s in the context of speech patterns and speech recognition and have more recently proved to be a successful tool in questions related to DNA sequence alignment. Mathematically, HMMs generalize classical discrete Markov chains, allowing for the possibility of switching between such chains based on a certain probability distribution. In this project we show how hidden Markov models can be used for identifying CpG islands. This question is important in gene prediction for identifying stretches of genomic DNA sequences that are biologically functional. Because CpG islands tend to appear near the promoters of important mammalian genes, HMM models that can identify CpG islands are valuable as gene finding methods (Durbin et al., 1998). In addition, abnormal methylation of the normally unmethylated CpG islands may be a pathway to cancer development (Jones and Takai, 2001), providing additional motivation to focus on methods for determining the locations of CpG islands.
AN EXAMPLE
We now present the module 1 in more detail. Due to the limited space, we have not included here a full description of the basic biological and mathematical background. Instead, we are providing only an outline of these components, focusing instead on one proposed pedagogical approach for using this module.
Biology Background
The module begins with an introduction on the need for and mechanisms of gene regulation. We then focus on the lactose (lac) operon, a gene regulatory mechanism that controls the transport and metabolism of lactose in
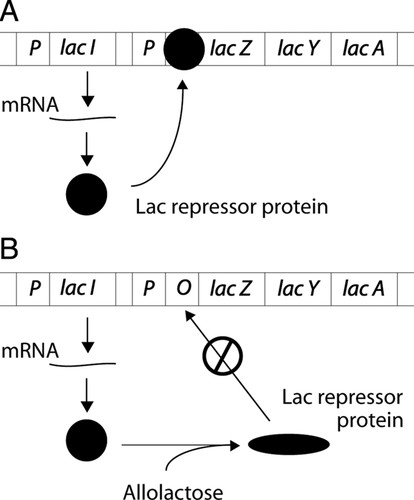
Figure 1. (A) lac repressor protein in action. The lac repressor protein binds the lac operon at the operator, preventing transcription of the lac operon mRNA. The operon is OFF. (B) Binding of allolactose to the lac repressor causes a conformational change in the repressor, preventing it from binding at the operator. Transcription of the lac operon mRNA can proceed. The operon is ON.
After the biological introduction, a class discussion is initiated to determine the state of the operon (ON or OFF) based on the presence or absence of external glucose and external lactose. This exercise has three main pedagogical goals: 1) to engage students in a discussion that reinforces the biological content, 2) to emphasize that the system is dynamic and its state changes with time as a result of interactions between its components, and 3) to guide students toward the realization that the discussion at this point is qualitative.
The dynamic character of the system is linked with the understanding that the state of a biological system at any given moment in time depends on the current configurations of the system's components as well as on their interactions. This is a fundamental prerequisite for understanding the dynamical nature of the mathematical models of the system. The qualitative character of the biological system should be linked with the understanding that the specific concentrations of lactose and glucose, as well as the exact concentrations of the proteins involved, are practically irrelevant. This justifies the need for a special type of mathematics appropriate in this situation. Unlike continuous (calculus-based) mathematics that uses all numbers on the real line representing concentrations and rates of change, discrete mathematics is more appropriate when only finitely many qualitative options are available; in this case two: present or absent, ON or OFF, 0 or 1, and so on. The appropriate mathematical concept is that of a Boolean variable. Boolean variables are allowed to take only two values, 0 or 1, representing numerically two mutually exclusive outcomes. When Boolean variables interact, they form Boolean networks.
Boolean Arithmetic and Dynamic Boolean Networks
We next introduce the main operations and arithmetic rules for Boolean variables: AND (denoted by the mathematical symbol ∧), OR (denoted by the mathematical symbol ∨), and NOT (denoted by the mathematical symbol ¬). In the context of Boolean networks we use the following intuitive definitions for the operations AND and OR: if two components, say x and y, of the system control a third component z, z = x∧y reflects the idea that x and y need to be simultaneously present (that is, have values 1) to affect z; z = x∨y represents the concept that x and y influence z independently and z is affected when either x OR y is present. Students are then asked to develop and discuss the tables of values for the operations AND, OR, and NOT, which leads to the introduction of the rules depicted in Table 1.
| Input | Output | |
|---|---|---|
| X | Y | Z = X∧Y |
| 1 | 1 | 1 |
| 1 | 0 | 0 |
| 0 | 1 | 0 |
| 0 | 0 | 0 |
| X | Y | Z = X∨Y |
| 1 | 1 | 1 |
| 1 | 0 | 1 |
| 0 | 1 | 1 |
| 0 | 0 | 0 |
| X | Z =¬X | |
| 1 | 0 | |
| 0 | 1 | |
A Boolean network is formed of interacting Boolean components, which are represented by Boolean variables. At this stage, the interactions are readily depicted in the form of a diagram, called a wiring diagram, that reflects the dependencies between the model components. Each square node in the diagram represents a component of the system, whereas links between nodes depict influential interactions: if x and y are two nodes of the graph, a directed link from x to y indicates that the quantity x affects the quantity y. Figure 2A presents a generic wiring diagram for a network composed of four Boolean variables denoted by x1, x2, x3, and x4.
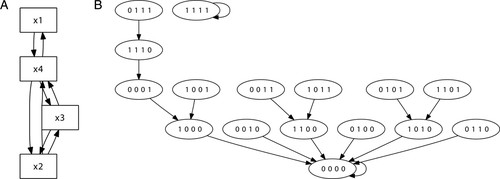
Figure 2. Wiring diagram (A) and the state space diagram (B) for the Boolean dynamical system in the example. Graphs produced with DVD (http://dvd.vbi.vt.edu).
Developing the wiring diagram of a Boolean network provides an excellent starting point for discussing conceptual generalizations, leading to the introduction of directed graphs (Figure 2A presents an example of a directed graph). In our experience, students readily accept this concept. To mathematics students, it is a generalization of regular graphs that occur very early in the mathematics curriculum. For biology students, directed graphs are very similar to the dependency cartoons that are commonly used to depict dependencies between interacting species in bimolecular networks or metabolic systems.
The next big step is to develop Boolean equations describing the specific interactions between the Boolean variables forming the system. Building on the previously discussed idea of dynamic changes, we introduce a transition function for each variable. The transition function fj describes how the variable xj will change in time under the influences (controls) of the other variables. Time is considered discrete; that is, changes to the system's state can only occur at fixed instances in time t = 1, 2, 3, …, beginning at time t = 0. Mathematically, the transition function fj of each variable xj is a Boolean expression of the variables influencing it. That is, if a directed link xi to xj appears in the wiring diagram, then the variable xi appears in the definition of the transition function fj. For example, the set of transition functions, 
The dynamic behavior of the network can now be computed from the Boolean equations above. Assume that at time t = 0, x1 = 0, x2 = 0, x3 = 1, x4 = 1. Then, at time t = 1, according to the transition functions above, we obtain 
In common with Figure 2A, Figure 2B also depicts a directed graph. This time, however, the directed graph represents the state transitions of the system. This shift in perspective provides another opportunity for a discussion focused on the information embedded in this graph. Discussion items include determining the identifying properties of the system's fixed points in terms of the directed graph of the system, examining the theoretic properties of the directed graph, understanding why, unlike the graphs of the wiring diagrams, there are no multiple edges between the vertices here, and discussing how the presence or absence of loops on the graph reflects the system's long-term properties.
For mathematics students, this discussion opens the doors to a class of mathematically challenging and, in some cases, open, problems. As mentioned above, when the number of Boolean variables in the network is large (and networks with hundreds of variables are of interest in biology), the number of states for the system grows exponentially with the number of variables, making the explicit computation of the entire state-transition diagram unfeasible. Still, questions about some of its essential properties may be possible to answer from the equations of the transition functions. Questions of importance include determining the existence of fixed points or limit cycles for the systems, determining the fixed points and limit cycles for a Boolean network, or (in case this is not possible) at least determining estimates for their number. The basic mathematics of Boolean modeling is accessible to practically anyone, and arises commonly in elementary discrete mathematics courses. The mathematics involved in the full project is appropriate for higher-level mathematics courses, as well as for student research projects. For example, when seeking to determine the system's fixed point, a mathematical reformulation of the transition functions as Boolean polynomials allows for the problem to be cast as one of solving systems of polynomial equations. In its general form, modeling and finding fixed points of the system can be pursued in the framework of polynomial algebra, specifically ideals over polynomial rings and their Groebner bases.
We have found that this fast track to a position where students can be introduced to high-level and unsolved mathematical problems is very exciting to them. As many of these questions represent an active area of research, students are enthused to engage with the problem and learn more about research efforts on Boolean networks by studying recent articles and results. This is in stark contrast to many students' experiences with calculus-based models, where one often needs several years of graduate work before one is able to understand the modeling methods currently used in journal publications. In addition, the tie between Boolean network models and the ability to gain a deeper understanding of the mechanisms of gene regulation serves as an important motivating factor. Although like any model, the Boolean approach captures only part of the underlying system, it offers a real innovation to tackle meaningful biological problems without the artificiality that often necessarily accompanies attempts to reduce calculus-based mathematical models to a student-friendly level, and in a context which students find refreshing compared with current canonical textbook examples, such as their repeated exposures to simple population growth models.
Boolean Network Models of the Lac Operon
The modeling process begins with identifying the major interactions and components of the lac operon system, as depicted in Figure 1, making choices of the model variables and parameters. The model variables are generally chosen to represent the major dynamic elements of the system (quantities that change with time), whereas the parameters correspond to static descriptors. We stress that different decisions regarding the exclusion or inclusion of any given component or part of the system will lead to different models. The next step is to define a wiring diagram for the model. Our first goal is to develop a Boolean model based on the minimal model approach for choosing variables and parameters used by Santillán et al. (2007).
After a guided discussion, students will likely identify the following elements as most essential to the lac operon regulation (the notation in the parentheses are the names we will be using for those elements from now on): mRNA (M), β-galactosidase (B), lactose permease (P), intracellular lactose (L), allolactose (A), external lactose (Le) and external glucose (Ge). Due to the fact that external conditions for the cell change slowly compared with the lifespan of E. coli, we can assume that Le and Ge remain relatively unchanged with time, assuming them to be constants and including them in the set of model parameters. The other quantities (M, B, P, L, and A) will be assumed to vary with time. However, it can be noticed that some of these variables exhibit related dynamics due to similarities in the underlying biochemical structures and mechanisms. Namely, because the structure of β-galactosidase is a homotetramer made up of four identical lacZ polypeptides and because the translation rate of the lacY transcript is assumed to be the same as the rate for the lacZ transcript, the number of independent model variables can be further reduced to three: M, L, and E, where E denotes the the lacZ polypeptide. This leads to a decision to use a Boolean network model with three variables—M, E, and L—and two model parameters—Le and Ge. The wiring diagram depicted in Figure 3 is developed next. Here, we have used an enhanced depiction of the links: a positive influence is indicated by an arrow; a negative influence is depicted by a circle.
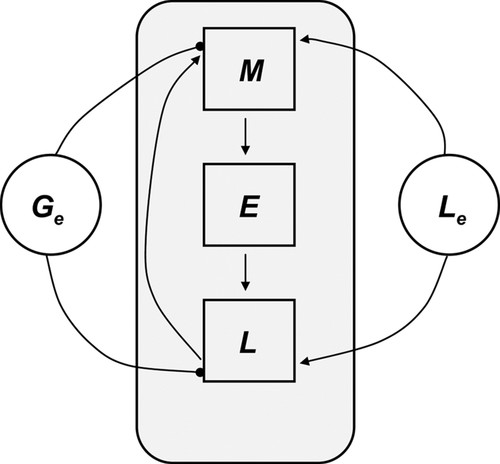
Figure 3. Wiring diagram for the minimal model. E denotes the lacZ polypeptide, M denotes the mRNA, and L denotes internal lactose. Le and Ge denote external lactose and glucose, respectively. The square nodes in the shaded rectangle represent model variables, whereas the round notes outside represent model parameters. Directed links represent influences between the variables: a positive influence is indicated by an arrow; a negative influence is depicted by a circle.
The transition functions for the variables M, E, and L are now specified from the wiring diagram and from the information regarding the biochemical interactions. We then develop the following model: 
Boolean Function for M. The first equation states that for mRNA to be present at time t +1, there should be no external glucose at time t, and either internal or external lactose should be present. Thus, when external glucose is present (Ge = 1), no mRNA will be produced (M = 0). Also, when there is no external glucose (Ge = 0) and there is lactose inside the cell (L = 1) or outside the cell (Le = 1), there will be at least a small number of lactose molecules inside the cell. This will cause mRNA production at time t +1.
Boolean Function for E. The production of mRNA (M = 1) will be followed by production of the lacZ polypeptide (E = 1).
Boolean Function for L. If there is no external glucose (Ge = 0), external lactose is available (L = 1), and permease (as represented by the polypeptide E) is present (E = 1), the permease will bring extracellular lactose inside the cell, ensuring the presence of intracellular lactose.
Analysis and Validation of the Model
Once a model is developed, it must be analyzed and validated. Students are cautioned that this is not an easy process and one must understand that a model can never be shown to be correct in an absolute sense. A model is just an approximation of the actual system and thus its validation is only appropriate within the context of the question that the model is developed to help answer. In our example, due to the simplicity of the model we are considering, it should be able to reflect only the basic qualitative properties of the lac operon. Thus, at a minimum, our model should show that the operon has two steady states, ON and OFF. When extracellular glucose is available, the operon should be OFF. When extracellular glucose is not present and extracellular lactose is, the operon must be ON. The module provides a series of exercises developed to demonstrate that the model satisfies these conditions.
We remind students that the operon is ON when mRNA is being produced (M = 1). When mRNA is present, the production of permease and β-galactosidase is also turned on. This corresponds to the fixed point state (M, E, L) = (1,1,1). In contrast, when mRNA is not made, the operon is OFF. This also means no production of either lactose permease or β-galactosidase. This corresponds to the fixed point state (M, E, L) = (0,0,0).
For our Boolean model of the lac operon, there are four possible combinations for the values Le and Ge of the model parameters: Le = 0, Ge = 0; Le = 0, Ge = 1; Le = 1, Ge = 0; and Le = 1, Ge = 1. For each one of these pairs of values, we compute the state space using the transition functions of the model. The results are shown in Figure 4. Notice that according to the model, the operon is ON only when external glucose is unavailable and external lactose is present. In all other cases, the operon is OFF, as should be expected. We think that this is a pedagogically important moment in the module as we get a chance to recall the results from our initial class discussion (based entirely on biochemical interactions) to determine the state of the operon (ON or OFF) based on the presence or absence of external glucose and external lactose. In this case, we have used a mathematical model that generates the same results.
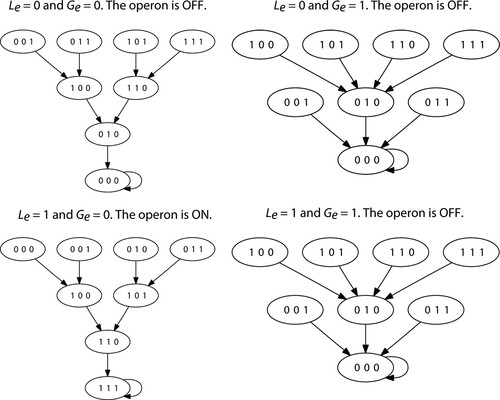
Figure 4. State space for the Boolean model of the lac operon for all possible combinations of parameter values. When external glucose is present, the operon is OFF. When external glucose is unavailable and external lactose is present, the operon is ON. Graphs obtained using DVD (http://dvd.vbi.vt.edu).
Further Exercises, Other Boolean Models, and Generalizations
At this point in the module, students should have acquired all of the mathematical tools needed to explore similar models on their own; consequently, we provide a series of exercises. One exercise presents the Boolean model developed in Stigler and Veliz-Cuba (2008) containing eight variables and accounting for the positive control mechanism of the lac operon regulated by the CAP-cAMP complex. Students are asked to 1) examine and comment on the wiring diagram of the model; 2) examine the model equations and compute a few model trajectories using the model transition functions; 3) use the DVD software to determine additional model trajectories and to compute the state-transition diagram for the model; 4) examine this diagram and determine whether the system has fixed points, limit cycles, or both; and 5) give biological justifications for the transition functions for each model variable. In another exercise, the students are presented with a flawed model of the lactose operon that seems to be legitimate. Students are asked to examine and validate the model and comment on the results. They should discover that the model produces one fixed point that is not biologically feasible, thus demonstrating a way in which validation attempts may fail.
The module ends with more challenging exercises referring to the bistability of the lac operon (Ozbudak et al., 2004), Boolean model approximations of dynamical systems, and developing Boolean models for the Lambda phage (Hinkelmann et al., 2009) with delay. As was the case when working with current research papers targeting mathematical problems stemming from the studies of Boolean networks, we found that students are excited to be engaged with these recent research studies and to apply their newly acquired expertise.
Suggested Use of the Module
We envision that this module can be used in biology courses such as genetics, microbiology, and cell and molecular biology and in mathematics courses in discrete mathematics, finite mathematics, mathematical modeling, and in advanced abstract algebra courses. The module is designed in a way that allows easy omission of any activities appropriate only for advanced mathematics or advanced biology courses without disrupting the general presentation. Instructors for lower-level mathematics and biology courses may choose to limit the use of mathematics to the use of Boolean algebra, directed graphs, and the time evolution of the simplest Boolean model described above. Instructors of higher-level courses may choose to also add some of the exercises using more advanced mathematics and/or biology (e.g., considering the mathematical questions outlined above or considering a more sophisticated model that incorporates the positive control mechanism of the lac operon). We expect that these choices will be affected by the primary designation of the course (biology or mathematics) in which the module is used, as well as on the personal preferences of the instructors. Respectively, the use of the module can span from one or two class meetings to up to 3 wk or more, especially if students are assigned all exercises and are required to research aspects of the current papers to which they pertain.
PRELIMINARY ASSESSMENT
Because our collection of modules is still under development, rigorous classroom testing and assessment results for the materials are not yet available. However, we have used parts of the modules as problems and projects in several mathematics and biology courses (from the elementary to the advanced level) including genetics, biomathematics, discrete mathematics, linear algebra, probability, geometry, algebraic geometry, and abstract algebra. This work on preliminary testing of the concept framework of the modules was useful in informing us about some qualitative specifics such as length of project discussions, total class times needed for the projects, balancing too-much versus too-little guidance by the instructor during the discussion, and deciding on the level of detail for presenting the biology and mathematics.
In addition, the content and pedagogical approaches for modules 1, 2, 3, and 4 were used by R. R. and T. H. during the minicourse The Mathematics of Systems Biology offered at the International Symposium on Biomathematics and Ecology: Research and Education (BERE-2009), Izmir University of Economics, Izmir, Turkey, June 13–17, 2009. This workshop provided an opportunity to present module-related material to an audience of faculty and students interested in connections between biology and mathematics. In anticipation of this workshop a survey was developed, then administered at the close of the workshop.
The survey consisted of 12 Likert-scale and four open-ended questions. Likert-scale options ranged from strongly disagree = 1 to strongly agree = 5, with neutral = 3. Additional questions asked for demographic data about the audience. Based on the 25 participants who completed the workshop survey, the data show respondents evenly split between university faculty (including a practicing M.D.) and students, with 80% designating their primary area of interest as mathematics. The audience showed a wide range of teaching experience that spanned many mathematics undergraduate courses ranging from calculus to linear algebra to mathematical modeling. Only one response indicated teaching experience in biology and several (n = 7) had no teaching experience (note: an open-ended question suggests that some of these were students looking for research or dissertation topics).
Overall survey results for the workshop were very positive. The Likert-item summary for those items related to workshop structure (five items), such as quality of activities and instruction, were all rated high with 90% of respondents agreeing or strongly agreeing at that level. The workshop consisted of three sessions and those were rated as 90, 85, and 88% agreement that the sessions were worthwhile. On two items that asked if participants deepened their understanding of biology and mathematics, they responded at 73% agreement for both, with 81% saying the workshop provided valuable professional development. Perhaps most relevant for this project is an item that asked if the workshop motivated the participant to try some of the [module] material in their courses, and 21% agreed and 58% strongly agreed that it did. One glowing response commented that the workshop was “one of the most useful presentations in my life.”
For evaluation purposes, this workshop survey represented a first attempt to systematically collect survey data that can directly inform us of faculty and student interest for using this type of materials and, possibly, recruit field-test candidates for later phases of the module development.
The planned structured evaluation activities for this project will consist of the following components and will be developed and coordinated under the direction of Dr. Steven W. Ziebarth, principal evaluator for this project: 1) A “teaching module” evaluation form will be developed to be administered after completion of each pilot and field testing of developed modules. Questions will be linked to both content and affective domains of interest for each specific module. Feedback for pilot phases will be summarized and given to authors for use in revising modules for further field testing. All data will be anonymous and only summary contents will be distributed to the authors by the lead evaluator. 2) A summary survey will be developed that gathers general affective information related to author perceptions and student attitudes that looks at usability across modules. 3) Further information on the modules will be gathered through selected interviews with authors and students (and other stakeholders as deemed warranted, e.g., instructors and/or colleagues in both the mathematics and biology departments) as needed to clarify issues arising from module evaluation data.
As with all curriculum development evaluations, some evaluation needs of this collection of modules will not be known a priori. Therefore, additional needed data will be collected as such circumstances warrant.
DISCUSSION
In this article, we present a collection of modules being developed by teams of mathematics and biology faculty from Sweet Briar College (SBC), a selective 4-yr liberal arts college for women; and Western Michigan University (WMU), a large regional university with Carnegie Doctoral/ High Research Activity status. At SBC, this ongoing project is the outgrowth of two successful National Science Foundation (NSF) curriculum development projects Division of Undergraduate Education (DUE) awards 0126740 and 0340930) for developing an interdisciplinary course in biomathematics and developing a textbook and laboratory manual for the course (Robeva et al., 2007a,b). For this project, we follow the PBL pedagogy adopted by R. R. and R. D. for the collection of modules they have developed for the laboratory manual (Robeva et al., 2007b) and draw on the lessons learned from the development, implementation, and assessment of this earlier work.
The modules are designed for use in a variety of courses outside of the traditional college calculus sequence. The modules are being developed as self-contained units that can be easily adopted by a broad spectrum of undergraduate institutions for use in courses ranging from general education mathematics, to teacher preparation, to advanced undergraduate courses. Moreover, one of our goals is to make the modules flexible: faculty looking for an exercise for a single course hour may choose only one part of a module, whereas faculty willing to commit more class time to these biomathematical applications can use the modules more extensively. We hope that students using the modules will experience higher levels of interest in their course work generated by the coupling of modern mathematics and modern biology, and an elevated awareness of biomathematics as an essential area of contemporary life. Likewise, we hope that creating and using the modules will encourage further cross-disciplinary faculty collaboration and development.
Because background in calculus is not required for most of the modules, and because significant parts of several modules require relatively low-level mathematics, these modules provide a particularly attractive entry into discussions of the need for mathematical models and of methods for building and using mathematical models. For example, constructing discrete models of biological networks requires only a modest amount of mathematical background and could be used as an introduction to mathematical modeling for students in the life sciences at an early point in their education. Such exposure could happen before they have taken calculus and calculus-based courses and have become familiar with differential equations and concepts that would allow for their analysis, including the study of directional fields, phase and time plots, null-clines, and bifurcation diagrams (Robeva and Laubenbacher, 2009). Moreover, as discrete models are only one step removed from the common way of depicting networks via wiring diagrams (often referred to as cartoons), they can serve as a natural, more rigorous, language in which to express the relationships among the molecular species in the diagram. Conversely, for mathematics students, discrete models of biological networks provide a meaningful way to introduce many of the concepts in the undergraduate curriculum, such as graphs, Boolean logic, polynomial algebra, dynamical systems, and mathematical modeling. Furthermore, these concepts can easily be connected with more advanced topics in the modern undergraduate discrete mathematics curriculum (Laubenbacher and Sturmfels, 2010).
Our materials are novel in the following ways. They 1) rest on mathematical underpinnings from topics and areas in modern discrete mathematics and algebraic statistics or new combinations of discrete and continuous methods; 2) focus on specific contemporary questions from biology with multiple, engaging facets; 3) arise from current scientific publications and ongoing investigations by leading researchers; 4) allow for parts of the module to be used in a progression of increasing difficulty from lower-level to higher-level mathematics courses; 5) utilize contemporary software (e.g., the DVD software used in the lac operon example) to make certain topics accessible at more elementary levels or suitable for inclusion in varying course frameworks by condensing or avoiding unnecessary computational details, thus allowing students to focus more fully on the interpretation of results; and 6) promote access to multilayered biomathematical materials across the mathematics curriculum, with the potential to increase student interest in topics both biological and mathematical.
Biology drives the presentation with the appropriate mathematical theory presented in the context of the problem-solving process. Most of the modules include hands-on exercises using relevant mathematical and statistical software. The mathematical content of the modules is appropriate for a variety of mathematics courses (in increasing level of difficulty) outside of the calculus-based sequences, including discrete mathematics, elementary linear algebra, elementary probability, applied matrix algebra, mathematical modeling, linear algebra, computational algebra and algebraic geometry, abstract algebra, and others. The early parts of the modules, paired with the description of the general mathematical background and the hands-on projects, also will be appropriate for use in existing biology courses including genetics, evolution, and cell and molecular biology.
Classroom testing for parts of the materials is currently planned or ongoing in selected courses at SBC and WMU. These include linear algebra, genetics, probability, biomathematics, and abstract algebra at SBC and a broad selection of courses at WMU catering to a diverse student audience including precalculus, discrete mathematics, elementary linear algebra, applied matrix algebra, numerical linear algebra, probability and statistics for elementary and middle school teachers, intro to modern algebra, survey of algebra, abstract algebra, discrete dynamical systems, and others. As soon as we finalize the initial drafts for each module, we will make these modules available for testing and adoption from www.biomath.sbc.edu at SBC and http://homepages.wmich.edu/∼hodge/at WMU3. When the drafts for the whole collection are finalized, our plan is to make the modules available nationwide through the MathDL (http://mathdl.maa.org/mathDL) and BEN (www.biosciednet.org/portal) portals of the National Sciences Digital Library.
In closing, we believe that our collection of modules will fill a niche and add useful resources to the existing literature of materials for undergraduate mathematical biology. Most importantly, by providing current, research-based applications for use throughout the mathematics and biology curriculum, we believe that this project will further increase student engagement and expertise at the critical nexus of molecular biology and mathematics.
FOOTNOTES
1 In what follows, the term calculus-based will be used to refer to mathematical techniques and models most commonly encountered in a traditional undergraduate calculus sequence. This includes Calculus I, II, III, along with a first course in differential equations, as well as their upper-division counterparts.
2 For a very simple example, the set V={(−1,0),(1,0)} consisting of two points in the xy-plane, given by the intersection of the parabola y = x2 – 1, and the circle x2 + y2 = 1, is exactly the set of common solutions of two equations whose terms are just polynomials in the variables x and y.
3 We expect to make the first drafts of modules 1, 2, 4, 5, and 6 available online by mid-August 2010 and the entire collection by the end of 2010.
ACKNOWLEDGMENTS
We thank Reinhard Laubenbacher (VBI) for generous assistance in identifying appropriate discrete models of the lac operon and recent publications of discrete models describing gene regulation and of reverse engineering methods. We also thank Steven Ziebarth (WMU) for summarizing the results and preparing the report for the postcourse assessment for the minicourse Robeva and Hodge offered at the symposium in Izmir, Turkey. We also gratefully acknowledge the support of the NSF under DUE award 0737467.



