RNA Secondary Structure Prediction by Using Discrete Mathematics: An Interdisciplinary Research Experience for Undergraduate Students
Abstract
The focus of this Research Experience for Undergraduates (REU) project was on RNA secondary structure prediction by using a lattice walk approach. The lattice walk approach is a combinatorial and computational biology method used to enumerate possible secondary structures and predict RNA secondary structure from RNA sequences. The method uses discrete mathematical techniques and identifies specified base pairs as parameters. The goal of the REU was to introduce upper-level undergraduate students to the principles and challenges of interdisciplinary research in molecular biology and discrete mathematics. At the beginning of the project, students from the biology and mathematics departments of a mid-sized university received instruction on the role of secondary structure in the function of eukaryotic RNAs and RNA viruses, RNA related to combinatorics, and the National Center for Biotechnology Information resources. The student research projects focused on RNA secondary structure prediction on a regulatory region of the yellow fever virus RNA genome and on an untranslated region of an mRNA of a gene associated with the neurological disorder epilepsy. At the end of the project, the REU students gave poster and oral presentations, and they submitted written final project reports to the program director. The outcome of the REU was that the students gained transferable knowledge and skills in bioinformatics and an awareness of the applications of discrete mathematics to biological research problems.
INTRODUCTION
Over the past several decades, biological research has expanded to include an extensive reliance on computational methods. Computational biology is routinely used to detect sequence similarities (homology) and to predict secondary structures as well as tertiary structures. Thus, computational methods are complementary to laboratory experimentation in molecular biology research.
Computational biology has experienced much growth over the past decade and has required mathematicians and biologists to collaborate to advance understanding in the biological sciences. Specifically, bioinformatics, which consists of applications of computational tools and approaches to analyzing biological data, has become crucial to research projects requiring molecular biology techniques. For example, methods and techniques for acquiring, storing, organizing, archiving, analyzing, and visualizing biological data require increased computational knowledge and skills. Thus, faculty from both biology and mathematics departments are encouraged to work on cross-disciplinary research projects and to create interdepartmental collaborations. Furthermore, to further advance research in computational biology, bioinformatics, and the biological sciences in general, there is much demand for collaboration among mathematicians, computer scientists, and biologists (Reed, 2004).
The terms bioinformatics and computational biology are often used interchangeably. However, bioinformatics more properly refers to the creation and advancement of algorithms, computational and statistical techniques, and theory to solve formal and practical problems derived from the management and analysis of biological data. Computational biology, in contrast, refers to hypothesis-driven investigation of a specific biological problem by using computers, carried out with experimental and simulated data, with the primary goal of discovery and the advancement of biological knowledge. Computational biology also includes lesser known but equally important subdisciplines such as computational biochemistry and computational biophysics. A common thread relative to bioinformatics and computational biology research is the use of mathematical tools to extract useful information from noisy data produced by high-throughput biological techniques.
Both bioinformatics and computational biology involve the use of techniques from computer science, informatics, mathematics, and statistics to solve biological problems. To facilitate the growth in biological research, which includes bioinformatics and computational biology, the authors in the BIO2010 report (National Research Council, 2003) discuss several measures that should be taken to equip biology students with skills and competencies needed to be effective biology researchers. One of the major recommendations of BIO2010 is to incorporate more computational classes into the biological curricula to prepare students for research careers, particularly in biomedical research.
In response to the needs of its biology students and the recommendations made in the BIO2010 report, Morgan State University (MSU), a mid-sized historically black university located in Baltimore, MD, has developed several programs, projects, and initiatives to prepare its students for careers that require interdisciplinary understanding, specifically an understanding of computational biology and bioinformatics. The Master of Science in Bioinformatics Program is a multidisciplinary program administrated by the Computer Science Department within the School of Computer, Mathematical, and Natural Sciences (SCMNS), which also houses the Departments of Biology, Chemistry, Mathematics, and Physics. The Bioinformatics Program fully integrates elements of computer science, mathematics, statistics, biology, chemistry, and physics into bioinformatics and computational biology. The objectives of this program are to offer students the theoretical foundations and practical skills in bioinformatics and to prepare students for careers in bioinformatics/computational biology within industry, academia, and government organizations. In addition, MSU has developed a Ph.D. Program in Bioenvironmental Science that is a didactic and research-driven program with participating faculty from departments within SCMNS. The program resides within the Department of Biology.
In addition to these programs, a Computational Biology course was designed and implemented to acquaint students with the concepts, principles, and methodologies of computational biology. The course consists of selected lectures in a multidisciplinary approach that covers topics in core areas of molecular and environmental biology, computer science, bioinformatics, and mathematics, in addition to hands-on computational laboratory exercises and problem sets. In addition, several research partnerships have developed between biologists and mathematicians that have served as the basis of several interdisciplinary initiatives at MSU. Interdisciplinary research teams conduct research in RNA structure prediction, which has experienced significant growth and advances over the past several years, primarily due to the availability of new experimental data and improved computational methodologies. Faculty in mathematics and biology are engaged in research that involves new methods for determining RNA secondary structures from sequence alignments, thermodynamics-based dynamic programming algorithms, genetic algorithms, and combined computational approaches. It has been reported in educational research literature that, based on experience of project-based teaching of bioinformatics within a cell biology course, the involvement of faculty with experience in pedagogy of different disciplines is recommended (Honts, 2003). Thus, the Research Experience for Undergraduates (REU) was conceived and designed around the topic of RNA secondary structure prediction and involved faculty with experience in pedagogy from the Departments of Mathematics and Biology at MSU.
RNA function is crucially dependent on secondary and tertiary structure but methods for prediction of structure from sequence require further development. Furthermore, under a suitable abstraction, RNA secondary structures can reveal surprising mathematical structure that is of interest to mathematicians. By their nature, biological sequences are often abstracted to discrete mathematical objects: strings over finite alphabets and their representation as trees and other graphs. This connection with combinatorics is particularly appropriate in the case of RNA secondary structures, and it yields a fruitful interaction between discrete mathematics and molecular biology. Thus, this topic is amenable to interdisciplinary teaching in the context of an REU.
To prepare students for programs and careers in bioinformatics and computational biology and provide undergraduate students with viable research experiences in these areas, the Department of Mathematics at Morgan State University received funding to create a summer REU program. The 6-wk summer program was designed to provide undergraduate students with a series of learning experiences aligned with the BIO2010 initiative and possibly increase their interest in several of the graduate-level programs offered at MSU. Thus, the purpose of this article is to highlight the MSU Mathematics Department 2009 REU Summer Program and some of the student outcomes from the project. We begin this paper by providing an overview of the REU program, the goals and objectives of the program, and the students who were targeted for the program. Next, we discuss the interdisciplinary content that was the primary focus of the program, and give an overview of the program activities and the nature of the student assessment used to evaluate the extent to which program goals and objectives were met. We conclude with the results of our assessments and a discussion of how the work of this project can inform current and future initiatives designed to reflect the goals and recommendations of BIO2010.
METHODS AND ASSESSMENTS
REU Program Overview
The 2009 MSU Mathematics REU was hosted by the Department of Mathematics at MSU during summer 2009. The REU was an activity of the Strengthening Undergraduate Minority Mathematics Achievement (SUMMA)/National Research for Undergraduates Program (NREUP) of the Mathematical Association of America (MAA) and funded by the National Science Foundation (NSF) and National Security Agency (NSA). The focus of the REU was to introduce undergraduate students to research centered on Riordan matrices with applications to RNA sequences from molecular and computational biology. In this REU program, two groups of undergraduate students studied certain RNA sequences related to viral and mammalian mRNA regulatory regions and used lattice walks modeled by Riordan matrices to predict RNA secondary structures.
Program Objectives
Current reform documents in both science and mathematics education provide strong philosophical support for the integration of science and mathematics as a way to enrich learning experiences and improve student understanding of and attitudes toward these disciplines (Berlin, 1989, 1991; Stepien et al., 1993; Berlin, 1994; Berlin and Hyonyong, 2005). The REU program objectives reflect these reform documents by focusing on improving students' knowledge, skills, and dispositions in mathematics and science by allowing students to make real-world connections between these disciplines. Specifically, students studied Riordan matrices and the related group properties in order to use lattice walks to predict RNA secondary structures. The following specific objectives were designed to achieve the goal of improving students' critical thinking, problem solving, presentation, and research skills.
It was expected that the students would:
Develop an understanding of discrete mathematics and its applications to biomedical research.
Develop a theoretical framework for understanding the function of RNA and for distinguishing between coding and noncoding regions of RNA.
Become acquainted with the structure and function of viral RNA genomes.
Learn the principles of base pairing in RNA and develop an appreciation of the role of RNA secondary structure in function.
Develop an understanding of the principles of thermodynamics underlying the folding of RNA.
Acquire skills of applying mathematics to modeling problems of biology, in this case RNA structure.
Develop skills for solving and manipulating generating functions and Riordan matrices in conjunction with proving Riordan matrix and group properties by using combinatorial and algebraic properties of the Riordan group.
Develop an understanding of the application of lattice walks to mathematical biology.
Enhance their scientific presentation skills.
Develop an interest in and motivation to pursue interdisciplinary research.
Student Qualifications
The REU program was designed for undergraduate students who had satisfied the following requirements:
Had completed at least 1 yr of university-level mathematics courses (e.g., Differential Equations, Linear Algebra, and Advanced Calculus)
Had an interest in conducting undergraduate research in mathematical biology
Had an interest in pursuing a graduate degree in the sciences and mathematics
Program Content
Biology Concepts: RNA, Structure, and Function. The REU students were instructed in the following background information in lecture settings and in group discussions with a faculty mentor. First, it was important for all students to clearly understand the structure of DNA in terms of the well-established Watson and Crick base-pairing rules. Secondary and tertiary structures of RNA predicate function, and because these rules also apply to RNA, with the substitution of U for T, it is possible to predict potential secondary structures from a linear sequence of RNA by identifying putative base-pairing partners (Leontis and Westhof, 2001). The rules for translation based on the genetic code also are largely universal and have been invaluable in gene predictions from the available genome sequences. Computational methods for detecting primary structure homologies and searching databases, nucleic acid, or protein alignments are available and are well developed (Jones, 2006); however, many challenges remain in the prediction of secondary and tertiary structures of biological macromolecules (Engelen and Tahi, 2010).
The rationale for developing computational methods for the prediction of secondary and tertiary structures of biopolymers emanates from the technical and cost limitations imposed by available biophysical methods. Like other biological macromolecules, the structure of RNA is determined with the biophysical methods of nuclear magnetic resonance (NMR) and x-ray crystallography. However, these techniques are expensive and not amenable to throughput analysis. They also require very specialized training of personnel. Thus, good algorithms for predicting RNA structure are promising approaches for gaining a better understanding of RNA function. The initial algorithms for predicting secondary structure in RNA sequences were developed with the assumption that the functionally important structures are the structures with minimal free energy and exploit empirical thermodynamics determinations (Eddy, 2004). One of the most widely used RNA folding programs, mfold, implements minimum free energy (mfe) algorithms that are mathematical models (Zuker, 2003). However, it is recognized that the functionally important structures are not necessarily the most thermodynamically stable structures because RNA molecules function in the context of protein complexes that may stabilize alternative structures by forming intermolecular contacts. Given that sequences that are functionally important tend to be more conserved, improvements on the original RNA folding approaches now incorporate sequence alignment in the secondary structure determination routine (Engelen and Tahi, 2010). Because secondary structure is more important for function in noncoding regions than the sequence per se, mutations that preserve secondary structure are more tolerated, and some RNA folding algorithms seek to capture covariation of two sites involved in base-pairing (Engelen and Tahi, 2010). Nonetheless, the procedures involve searching for alternative secondary structures and then determining the species with higher probabilities of being biologically relevant.
The second level of instruction was intended to convey the importance of structure to function of RNA. It was emphasized to the students that secondary and higher-order structures play an important role in the function of RNA molecules and can be summarized as follows. The secondary structure of the upstream and downstream untranslated regions of mRNA, the 5′-untranslated region (UTR) and 3′-UTR, is very important in regulating translational efficiency (Gray et al., 1998). In addition to the more abundant species of RNA—mRNA, rRNA, and tRNA—that are involved directly in protein expression, it is now recognized that other minor species of RNA, originally collectively termed as small nuclear RNA and small nucleolar RNA, are intricately involved in RNA processing and maturation and in the regulation of translation. A second class of small nuclear RNAs called microRNAs is expressed in most eukaryotes and also regulates mRNA translation. The microRNAs are transcribed as larger molecules termed as primary microRNA (pri-miRNA) and processed in the nucleus into intermediates (pre-miRNA) containing a stem-loop structure that are exported to the cytoplasm for processing and integration into the mRNA-degrading enzyme Dicer (Mendes et al., 2009). In fact, according to Tamar Schlick, microRNA molecules are integral components of the cell machinery for protein synthesis, transport, editing, chromosome replication and regulation, and catalysis, among many other functions (Schlick, 2006).
The RNA genomes of different classes of viruses are characterized by highly conserved regions within the untranslated 5′ and 3′ regions. These regions mediate such diverse functions as selecting the site of initiation of translation through internal ribosome entry, control of genome packaging, and control of replication and transcription (Harris et al., 2006). These regions by and large function through formation of secondary structures, consisting of stem-loop structures that may or may not have one or more bulges or mismatches, and pseudoknots, which are higher-order structures comprising two overlapping stem-loop structures (Leathers et al., 1993).
In the REU, one group of students chose to model functionally important sequences of yellow fever (YF) virus after a literature search and based on group members' interest. YF virus is a Flavirus that is transmitted by mosquitoes, and it is re-emerging as an important human pathogen in most parts of Africa and South America (Bryant et al., 2007). The genomes of Flavirus and other plus strand RNA viruses are synthesized from minus strand templates, and the 3′-UTRs therefore contain information for replication of the viral genome (Yu and Markoff, 2005). Stem-loop structures in this region are essential for replication competence, and secondary structure patterns of the 3′-UTR of the YF virus have been correlated to virulence, with vaccine strains exhibiting distinctly different folding patterns to those of infectious strains (Proutski et al., 1997). The 5′-UTR of Flavivirus genome also exhibits potential to form stable secondary structure elements that are essential for translation of viral genes (Chiu et al., 2005). Consistent with the viral replication cycle whereby the plus strand genome is synthesized from a minus strand, mutations in the 5′-UTR decrease replication perhaps due to defective synthesis of the template minus strand (Yu et al., 2008).
After the literature search and consultations with faculty, one group of students decided to use the 3′-UTR of YF virus as the subject of structure analysis. The second group focused on uncharacterized messenger RNAs that are associated with epilepsy.
Discrete Mathematics Concepts
It has been reported in the education literature that although most students will have been exposed to discrete mathematics at different times during their education, it is clear that the carryover to applications in other disciplines is limited, and it is proposed that other methods for teaching quantitative skills should be developed (Labov et al., 2010). The mathematical topics, ideas, techniques, and tasks that were expected of the students to accomplish the goals of the REU project included the following: 1) solving and manipulating generating functions and Riordan matrices in conjunction with proving Riordan matrix and group properties by using combinatorial and algebraic properties of the Riordan group, and 2) studying lattice walks and applications to mathematical biology.
Riordan matrices are a special subset of infinite ordered lower-triangular matrices. The set of all Riordan matrices forms a noncommutative group call the Riordan group (Shapiro et al., 1991). The Riordan group is formed under matrix multiplication. The famous Pascal triangle written in lower-triangular form is a typical example of a Riordan matrix and thus an element of the Riordan group. The students specifically studied two other elements of the group that are Riordan matrices we call RNA matrices (see Figure 1). The entries in the first column of the RNA matrices count the RNA numbers. These numbers have applications to lattice walks and RNA sequence prediction.
The students required some basic background information from matrix algebra and combinatorial analysis. When constructing Riordan matrices, emphasis is placed on basic concepts related to generating functions (or formal power series). The notion of a generating function is a formal process that provides an efficient way to represent an entire sequence associated with a formal power series. The students needed to understand basic concepts of generating functions. Thus, in reference to generating functions, the students studied properties with emphasis on convolution (Cauchy) products, closed form representations, composition of generating functions, and derivation of generating functions from recurrence relations. Given this background information, the students were able to define the RNA matrices mentioned above. The basic biology needed to understand RNA primary sequences was also introduced during the program.
In addition to understanding the above-mentioned ideas, students were exposed to the concept of lattice walks. A lattice walk is a unit-step path that moves from one point to another such that unit steps are allowed to move in a discrete number of directions such as up denoted by N (north), down denoted by S (south), and right denoted by E (east). The walks are in the first quadrant of the (x, y) plane, and two types of lattice walks are considered. First, we consider lattice walks that are of length n and height k that start at the origin (0, 0) and never go below the x-axis such that there are no consecutive up and down (NS) steps. Then, we consider lattice walks that are of length n and height k that start at the origin (0, 0) and never go below the x-axis such that there are no consecutive down and up (SN) steps.
There is a one-to-one correspondence between the set of lattice walks of length n as defined above and RNA secondary structures of length n (where n is the number of bases) (Nkwanta, 1997, 2008). The correspondence is set up by the following rules. We represent a secondary structure sequence of length n as nonintersecting arcs (or chords) along a horizontal axis. The RNA sequence is then represented by a sequence of integer points increasing in order from left to right along the horizontal axis where base pairs (or bonds) are denoted by arcs and unpaired bases are denoted by nonarcs. We then consider along the horizontal axis whether two integers points are paired by an arc at particular positions i and j or whether an integer point is unpaired by a nonarc at a particular position k. We then consider the positions i and j along the horizontal axis that are paired as a set of integer pairs {(i, j) 1 ≤ i < j ≤ n}. The pair (i, j) indicates a bond between the bases at positions i and j of the corresponding strand and are represented by N and S steps of a lattice walk. An unpaired base at position k of the corresponding strand is represented as an E (east) step. So, i is associated with N steps, j with S steps, and k with E steps. For example, consider the primary sequence ACUACGU of length n = 7 bases. We consider the sequence as a folded structure where base A is paired at position 1 with base U at position 7 and base C at position 2 is paired with base G at position 6 as nonintersecting arcs along a horizontal axis. The one-to-one correspondence is defined where the bases corresponding to the integer pairs (1, 7) and (2, 6) are associated with (N, S) steps relative to the corresponding walk, and all other bases corresponding to the integers are associated with E steps. Thus, the corresponding lattice walk is NNEEESS.
The number of different lattice walks of length n and ending height k (n, k ≥ 0) as defined in the previous paragraph are recorded by the n by k lower triangular arrays (Figure 1).
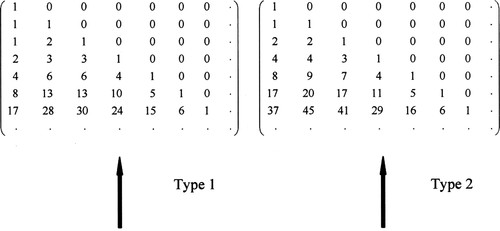
Figure 1. Triangular arrays.
In the leftmost column of both arrays lies a sequence of numbers known as the RNA numbers. These numbers, in addition to counting lattice walks, also count the number of possible RNA secondary structures of nucleotide length n.
Program Activities
Before the 6-wk on-campus session, the five students were introduced to basic biological concepts on RNA secondary structure prediction and mathematical concepts on combinatorial structures called lattice walks. This 2-h pre-REU session was held 1 mo before the start of the summer program and was designed to familiarize students with the basic mathematical and biological concepts related to the REU summer project. Students were given a brief overview of the connection between RNA primary sequences and a certain subset of lattice walks. At the end of the 2-h session, students were given reading materials that were relevant to the REU and reflected possible research topics for their projects.
The first 2 d of the REU, the students, as a team, were required to study the fundamental theorems of arithmetic, algebra, and calculus and give a brief PowerPoint presentation of their understanding of the theorems. This activity was designed to encourage students to complete a task in a short time, to promote teamwork, and to familiarize them with giving mathematical presentations.
After the team-building activity, the students were given introductory lectures on mathematical biology involving the connection between lattice walks and RNA secondary structure. Given a primary RNA sequence of a known RNA secondary structure, the students were taught how to determine whether a stable RNA secondary structure can be predicted (or modeled) by using lattice walks modeled by Riordan matrices.
A major activity of this REU project was having students complete research projects in teams. The five undergraduate students were divided into two groups to complete projects. One group's project focused on a certain RNA sequence related to the YF virus, and the other group's project focused on a certain RNA sequence related to epilepsy. The results obtained by the students were that they established direct links between lattice walks and primary RNA sequences of YF and epilepsy and used lattice walks modeled by Riordan matrices to predict more stable RNA secondary sequences related to these two diseases. In addition, one student was able to obtain a mathematical result by proving a new recursion for the RNA. The objectives of these projects were to help improve the critical-thinking, problem-solving, and research skills of the student participants.
Finally, because one of the major goals of this project was to help improve the critical-thinking, problem-solving, and research skills of the student participants, the students also were required to conduct library and Internet database searches and give several written and oral presentations throughout the program. They also were exposed to mathematical software like Maple and Scientific WorkPlace, the biological database BLAST, and the RNA sequence prediction program mfold.
Project Timeline
First week, sessions 1 and 2, 9:00–12:00 and 1:00–4:00 (Riordan matrices) | |||||
Second week, sessions 3 and 4, 9:00–12:00 and 1:00–4:00 (Riordan group) | |||||
Third week, sessions 5 and 6, 9:00–12:00 and 1:00–4:00 (lattice walks and RNA) | |||||
Fourth week, sessions 7 and 8, 9:00–12:00 and 1:00–4:00 (RNA prediction of viral RNA) | |||||
Fifth week, sessions 9 and 10, 9:00–12:00 and 1:00–4:00 (continue viral RNA application) | |||||
Sixth week, sessions 11 and 12, 9:00–12:00 and 1:00–4:00 (papers/reports due) | |||||
Student Assessment and Program Evaluation
The project team developed a series of assessment activities designed to measure the effectiveness of the project and determine the extent to which students' knowledge, skills, and dispositions changed as a result of their participation in the REU project. To assess the extent to which students' knowledge and skills in the REU content areas had progressed over the course of the project, the project team implemented several formative and summative assessments. First, students were required to submit weekly written reports that reflected the mathematics and biology content discussed during the week. These reports were evaluated by the team based on the content objectives. Second, students' weekly informal presentations of relevant biology and mathematical concepts were assessed to determine how students' knowledge of the material was evolving throughout the program. Third, after 2 wk in the project, students created posters and participated in a research poster session. These posters were assessed using an evaluation rubric that reflected content goals, presentation skills, and research skills that the project team wanted students to learn. As summative assessments, students' final oral presentations and final written reports were evaluated to determine whether or not students learned the concepts reflected in the program objectives.
To assess the changes in students' dispositions over the course of the project, several assessments were used. First, informal observations of REU sessions were used to determine students' attitudes and interest in the project and how these evolved during the project. Second, the project team conducted informal interviews with students where various questions were asked to understand the students' experiences of the project, particularly how their attitudes and interest in the project were evolving over the course of the 6 wk. Third, student attendance was recorded to assess their commitment to the project. Finally, at the end of the REU students were given exit interviews that revealed students' conceptual understanding of the content, their attitudes toward the program, and their interest in continuing their REU participation and research in the future.
RESULTS
The evaluation of the REU project was determined by analyzing the above-mentioned assessment data to evaluate the extent to which all of the expressed project goals and objectives were met. Based on the student assessment data, the REU project had a positive impact on all participating students. This positive impact was demonstrated in several ways. First, weekly and final reports revealed that the students developed a deep conceptual understanding of integrated mathematics and biology concepts, particularly as they related to RNA and lattice walks.
In addition to increased understanding of interdisciplinary content, the REU students also demonstrated positive attitudes toward interdisciplinary content and research. Last, several students developed their own research that extended the work of the project and presented their work at several conferences. To capture the impact of the project, students were asked to share their experience of the program and document what they learned as a result of their participation. Some of the students' comments regarding their experience in the program follow:
Student A: “The project covered a lot in terms of research especially in Biology. I was able to learn a lot about biological research and my understanding of Maple was solidified. I was also introduced to mfold. My research skills were greatly improved as a result of this project. This project mainly improved my research skills, which were minimal at best before the project. Also, the mathematical recursion that we came up with tested my persistence, as I tried to prove it but could not find a quick and easy solution. These two factors have made me aware that research takes more than an idea, and that one has to be really dedicated to the work in order to get any tangible results.”
Student B: “As a result of the REU, I can view the world and my education in a different way now that I have learned methods to apply math and science. I have matured in these areas because my level of understanding has transitioned from being only taught what a teacher tells me to learning and furthering my knowledge with research and applications. Before the REU, I did not think as much for myself. Critical-thinking, problem-solving, and research skills are very necessary to further yourself in higher education. It has helped me focus specifically on a specific problem and concentrate on learning more that could help with the particular problem such as how the problem occurred, what does the problem do, and what are possible solutions.”
Student C: “The REU introduced me to the combined methods of using biology and mathematics. Being as though it was my first experience using the two intertwined, I would have to say that my maturity was broadened. I came into the REU with limited understanding of the connection between math and molecular biology. On completion of the REU, I gained a greater appreciation of the work and a curiosity that is leading me to want to further the research.”
Student D: “We covered a wide range of mathematical and biological topics, which was sometimes intimidating but insightful nonetheless. Although I took some time for me to get comfortable enough with the project to decide a logical course of action, it taught me to be practical with my research goals. Aside from classroom projects and papers, this was my first experience on my own research project. The entire process of highlighting a problem by asking fundamental questions, then devising a method of attack using a computational algorithm to finally implementing it has built my confidence and I really would like to do further research in the future probably as a career, too.”
Student E: “The REU helped me to identify some of my weaknesses with regard to working in groups, like communication of ideas, and tolerance of differences. This experience has motivated me to pursue a course in Group Dynamics. My understanding of the connection between math and biology as well as my interest in continuing research in this field was improved.”
In addition to sharing these experiences, REU students presented their summer projects at the MSU Summer Research Symposium. This is a summer symposium for all summer research programs in the School of Computer, Mathematical, and Natural Sciences. The students also were required to give a presentation in the Department of Mathematics at the end of the summer program and to submit a final paper. The summer research projects are summarized below.
Title 1: A Study on RNA Secondary Structure Prediction of the Yellow Fever Virus Using a Lattice Walk Approach
ABSTRACT: In this research we hypothesize that a direct link is established between lattice walks and the RNA sequence of a short domain of the 3′-UTR of a selected strain of the YF virus and a more stable strain is predicted. The 3′-UTR is important for genomic packaging and knowledge and manipulation of the folding patterns of its secondary structure can potentially affect important viral functions like translation and replication. Motivation for study of the YF virus is due to its resurgence in Africa, South America, the Caribbean, and other parts of the world, and its inclusion among the list of potential bioterrorist weapons in the United States. We use a lattice walk approach that involves analyzing relationships between lattice walks and RNA sequences, applying enumerative combinatorics on the number of possible RNA sequences, and evaluation of the mfe of strains of RNA sequences identified with lattice walks. As with other mathematical models, the lattice walk approach allows for comparison of linear (primary) sequences, but it also provides the flexibility of identifying and comparing possible secondary structures for a given primary sequence. Using a 20-nucleotide sequence of the 3′-UTR of the Trinidad 79 strain, combinatorial formulae were used to count the total number of possible secondary structures that could result. These formulae were also used to reduce the number of counted secondary structures. The lattice walk approach was used to identify possible arrangements of the primary sequence for prediction of secondary structure. Analyzing lattice walks linked to the primary sequence and running an application of mfold (an RNA and DNA computerized folding package) allowed for the design of new structures. Minimum free energy calculations generated by mfold were used to reveal the most thermodynamic conformation. Application of varying lattice walks to the original primary sequence showed that the most stable structure was equivalent to that originally generated by mfold, with an mfe reading of –9.9 kcal/mol. When the primary sequence was manipulated by mfold, lattice walks generated even more stable structures; the most stable of which had an mfe reading of –16.3 kcal/mol. The results clearly indicated that a direct link was established between lattice walks and RNA sequences of the 3′UTR of the Trinidad 79 strain, and a more stable secondary structure was predicted. These predicted secondary structures require more biological testing and further experimental and laboratory research to determine biological functionality and application to the YF virus. Establishing the biological function of the predicted structures may aid in both antiviral drug design and vaccine development, as well as provide protection against the threat of YF as a bioterrorist weapon, and help YF researchers better understand the control of the spread of the virus.
Title 2: Lattice Path Model of Epilepsy Type 2 (EPM2A): mRNA Secondary Structure
ABSTRACT: This research attempts to develop an mRNA structure or motif marker for epilepsy. The thermodynamic algorithm mfold is used to predict the most stable secondary motif for eight single-stranded mRNAs implicated in EPM2A epilepsy that were then compared, and preliminary results indicated that there are conserved, similar secondary motifs in all the RNA's analyzed. As such, the hypothesis is that these conserved and similar motifs are indicative of the phenotype epilepsy and can be modeled by using lattice paths to represent other forms of epilepsy.
Finally, REU students showcased their research by presenting at several conferences, including the National Association of Mathematicians annual conference, the Annual Biomedical Research Conference for Minority Students, and the Joint Mathematics Meetings Research. Presentations included the following: A New Recursion for the RNA Numbers in the RNA Type II Array, RNA Secondary Structure Prediction, Lattice Walks and the Yellow Fever Virus and RNA Secondary Structure Prediction, and Lattice Walks and the Yellow Fever Virus. The YF project received a student award for their poster at the 2010 Joint Mathematics meetings.
DISCUSSION
Among the objectives for this project were to 1) help students develop a theoretical understanding of the structure and function of RNA and the role of untranslated regions in the viral genomes; 2) cultivate an appreciation of the role of secondary structure in RNA function; 3) acquire skills of applying mathematics to modeling problems of biology, in this case RNA structure; and 4) help improve the undergraduate students' critical-thinking, problem-solving, and research skills. The results of this project suggest that students did meet these goals and felt that the experiences they gained through the project were useful and helped foster an understanding of and appreciation for RNA research. For example, after the initial training in theoretical considerations of RNA secondary structure and function, the students concluded primary literature searches on the role of RNA secondary structure in viral replication and identified the YF virus as a pathogen of interest based on the biomedical needs such as the absence of efficacious vaccines. This led them to investigate a region of the RNA genome that shows variability in structure between different strains of the virus. The lattice walk was applied to a sequence within this region that is proposed to form a long stable hairpin structure and is located within the 3′-UTR of the YF genome. The most stable structure, based on the length and composition of the stem, was identical to one obtained with a genetic algorithm combined with sequence analysis (Proutski et al., 1997). The same results also were obtained when the region was analyzed with mfold, a widely used nucleic acid folding program (Zuker, 2003), indicating the validity of using lattice walks in RNA folding problems. The students therefore applied mathematical principles to a biological problem and then validated their results with an established program, thereby giving them an insight into the mathematical basis of bioinformatics tools that are now easily available on the Internet.
The various tests performed revealed certain implications, namely, that the configuration of the walks of the RNA sequence might be key to identifying structure with the minimum free energy. For example, the students observed that an increase in the height of the walk, indicating the length of the stem, could lead to a reduction in the minimum free energy for sequences with the same nucleotide composition.
The results of this project were particularly compelling given that the students targeted for this project were not necessarily students with high grade point averages, nor were they initially interested in interdisciplinary research. A rationale for targeting students with a wide range of academic profiles was to encourage several types of students to engage in interdisciplinary learning and research experiences that potentially would motivate them to pursue academic programs and careers in these fields. Students reported that they found these experiences very valuable, and many of them expressed an interest in expanding the work that they had begun in the REU summer institute. Furthermore, these students' understanding in both mathematics and biology were enhanced as a result of their participation, which could potentially have implications for their academic achievement. This suggests that students of all academic levels can benefit from such a summer program, and the recommendations proposed by BIO2010 can not only serve high-achieving students but could potentially impact students with a variety of academic profiles. Specifically, the recommendation to encourage students to engage in independent research is appropriate for all students. Hence, programs developed to engage students in this type of research should recruit students with a variety of academic profiles.
Also, the results of this project suggest the importance of giving students the opportunity to present their work to the broader scientific and educational community. Four of the five students who participated in this program presented their work at various local and national conferences. They admitted that presenting their work was extremely valuable because they had never before attended national biology, mathematics, or both conferences so they developed a new insight into what it meant to present scholarly work. This exposure to the broader scientific community was critical for the students' sense of themselves as budding scholars and researchers. Hence, it is important that similar projects build into their program opportunities for students to showcase their work to broader audiences, which will have lasting impact on fostering the next generation of biological researchers. Through this active-learning experience, students of various science backgrounds were able to come together and apply what they learned in the classroom, particularly the combinatorics of RNA, to solve a very concrete problem in computational biology. Hopefully, as more and more of this type of collaboration develops, it will lead to a greater awareness of the benefits to be gained from combining the life sciences with the computational sciences.
In addition to the positive outcomes of the project, typically there are several challenges encountered in the collaboration between biologists and mathematicians. One of these challenges is how to bridge the different approaches to research between biologists and mathematicians. Most biologists are trained to carry out the experiment directly in a laboratory without engaging in rigorous mathematical concepts. Mathematicians, in contrast, tend to develop theoretical solutions that are not readily understandable to biologists. Another challenge lies in the disparate and often highly technical use of terminology, symbols, and acronyms in both fields. Some life scientists are not always able to understand and interpret certain mathematical languages and interpretations of mathematical findings. Likewise, some mathematicians may find it difficult to understand how life scientists present and interpret their data and findings. The type of collaboration described in this REU serves the additional purpose of bridging such gaps, as the students were required to form small cohesive teams and to work to solve the problems jointly, irrespective of their backgrounds.
ACKNOWLEDGMENTS
We acknowledge the SUMMA/NREUP of the MAA, the NSF (grant DMS-0552763), and the NSA (grant H98230-06-1-0156) for financial support for the REU. We also acknowledge students Kemi Adeyinka, Ashley Banks, Damond Collier, Anya Ecto-Joseph, and Daudi Sagalla, who participated in the REU. We thank them for participating. We also thank Julian Fuller, the REU graduate student assistant for his assistance. James Wachira was supported by the Maryland Technology Development Corporation and Army Medical Research and Material Command (grant W81XWH-07-2-0055).



