A Summer Program Designed to Educate College Students for Careers in Bioinformatics
Abstract
A summer program was created for undergraduates and graduate students that teaches bioinformatics concepts, offers skills in professional development, and provides research opportunities in academic and industrial institutions. We estimate that 34 of 38 graduates (89%) are in a career trajectory that will use bioinformatics. Evidence from open-ended research mentor and student survey responses, student exit interview responses, and research mentor exit interview/survey responses identified skills and knowledge from the fields of computer science, biology, and mathematics that are critical for students considering bioinformatics research. Programming knowledge and general computer skills were essential to success on bioinformatics research projects. General mathematics skills obtained through current undergraduate natural sciences programs were adequate for the research projects, although knowledge of probability and statistics should be strengthened. Biology knowledge obtained through the didactic phase of the program and prior undergraduate education was adequate, but advanced or specific knowledge could help students progress on research projects. The curriculum and assessment instruments developed for this program are available for adoption by other bioinformatics programs at http://www.calstatela.edu/SoCalBSI.
INTRODUCTION
Sequencing the human genome fueled the creation of tools to analyze data generated by this international project and opened the door to the widespread development of bioinformatics as an academic discipline. In broad terms, bioinformatics explores the interface of biology and computers (Pevsner, 2003). A narrower definition of bioinformatics, and the definition used in this article, is the use and development of computer databases and computer algorithms to analyze molecular biology data. Many formal academic programs in bioinformatics became available to students shortly after the sequence of the human genome was published in 2001 (Lander et al., 2001; Venter et al., 2001), and since that time, bioinformatics programs have been developed that range from undergraduate certificate programs to doctorate degree-granting programs.
As is typical of any new discipline, the subject content in current bioinformatics academic programs is not identical. For example, in six Australian universities offering bioinformatics undergraduate degrees, the percentage of content devoted to bioinformatics, mathematics, statistics, computer science, and biological sciences ranged widely (Cattley, 2004). Preparing students for careers in bioinformatics and providing them with experiences upon which to make a career choice are particularly challenging for colleges and universities that do not have concentrated programs in bioinformatics or bona fide bioinformaticists at their facilities. Since 2003, California State Los Angeles University (CSULA), an institution with no faculty formally trained in bioinformatics, has operated an intensive summer bioinformatics education program emphasizing didactic training, research training, and professional development. The curriculum is designed to educate upper-division undergraduates and first- and second-year graduate students from a wide range of backgrounds (including but not limited to biology, computer science, and bioinformatics). The program, named Southern California Bioinformatics Summer Institute (SoCalBSI) is one of nine Bioengineering and Bioinformatics Summer Institutes (BBSIs) participating in a National Institutes of Health–National Science Foundation joint program to train students in bioinformatics and/or bioengineering. Each BBSI has a didactic and research component, but they differ in content and curriculum design (Munshi et al., 2006). Because SoCalBSI is unique among the BBSIs in that it provides students offsite internships, it is particularly well suited for collecting feedback on its curriculum from a broad range of professional bioinformaticists (research mentors) both in academia and industry.
In this article, we offer qualitative evidence (open-ended research mentor and student survey responses, student exit interview responses, and research mentor exit interview/survey responses) derived from our experiences in this program that identify skills and knowledge that are critical for students considering a career in bioinformatics. This article also describes the structure of our 10-wk program and the assessment strategy used to ensure its continuous improvement based on feedback from students and research mentors. These data can be used to inform the development of formal curricula that prepare students for successful research experiences in bioinformatics, especially at schools lacking intensive bioinformatics research programs.
An ancillary focus of this article is to provide qualitative assessment instruments and procedures that can be easily adopted by other bioinformatics programs. These instruments include 1) online didactic instructor evaluation forms that address general aspects of instructor performance as well as attainment of objectives specific to bioinformatics curricula, 2) student and research mentor open-ended surveys that address curriculum and work ethic, 3) student exit interview questions that address all aspects of the program, and 4) research mentor exit survey or interview questions that also address all aspects of the program.
PROGRAM DESIGN
SoCalBSI was funded through a grant sponsored by the National Science Foundation and the National Institutes of Health (http://bbsi.eeicom.com). The objective was to train upper-division undergraduates and first- and second-year graduate students for careers in bioinformatics. The 10-wk program was divided into two phases. The first phase consisted of 3 weeks of didactic instruction in bioinformatics, molecular life science, computer science, bioethics, and mathematics. In the second phase, students performed bioinformatics research under research mentors at one of nine institutes in the southern California area. Students also received professional development training on Fridays.
Student Selection
Bearing in mind that the goal of the program is to prepare students for entry into the bioinformatics workforce, we sought academically motivated students who possessed an education background that demonstrated an interest in bioinformatics. We held the belief that bioinformaticists should be trained to use existing popular bioinformatics software programs, and, in addition, possess the capability to develop applicable software programs that will advance the field. Given this philosophy, students were required to have completed at least one course in molecular life science and one course in computer programming before applying to SoCalBSI.
SoCalBSI brought together students with different academic backgrounds from across the United States. The mix of academic levels provided an opportunity for undergraduates to learn about graduate school experiences from the graduate students. Students were required to have achieved a minimum 3.0 GPA to enter the program to ensure that they would be able to keep pace with the didactic session. Each applicant was asked to provide one letter of recommendation from an individual who could judge the academic or research quality of the student and a second letter of recommendation from a personal reference. Students were asked to explain why they wanted to join the program, to give a description of previous research experience, and to provide grade transcripts, which were used to verify academic records and ensure that students met the minimum course requirements.
Students who met the eligibility requirements were selected into the program on the basis of five criteria: 1) academic success, 2) research experience, 3) appropriateness of educational background, 4) fit for our program, and 5) potential to increase workforce diversity. Applications were available online starting January 1. We found that an early March deadline for application submission was important to successfully compete with other summer research programs with similar commitment deadlines.
From 2003 through 2005, there were 155 applicants of whom 38 joined and completed the program. The program did not keep track of the number of offers to applicants that were turned down. The average GPA of the students in the program was 3.5. The numbers of students enrolled each summer were 14, 13 (including 2 students who returned from the previous year), and 13. Students were paid biweekly from a stipend of $6000 for undergraduates and $7000 for graduates. They were given the option of student housing on campus and the cost for housing was deducted from the stipend.
Who Applied to the Program?
To better understand the experience of students interested in bioinformatics, we present an overview of the education background of the students who applied to our program. Applicants to our program were enrolled in science majors at their home institutions and could be grouped into one of four categories: computer science, molecular life science, “blend,” and “other” (hereafter, these categories are referred to as blend and other, respectively). Students belonging to the computer science category were computer science majors and computer engineering majors, and they rarely had more than one course in molecular life science. Students in the molecular life science category majored in biology, molecular biology, biochemistry, and biomedical engineering, and they usually had not completed more than one computer programming course. The blend majors included cybernetics, bioinformatics, computer science with an emphasis on bioinformatics, and double majors in molecular life science and computer science. These students had equally strong backgrounds in molecular life science and computer science. The other category included students studying a variety of fields outside of the aforementioned categories, most frequently mathematics.
Figure 1 shows the number of students who applied and were accepted into the program, stratified by year and major category. Out of the applicant pool, 50 students were majors in molecular life science, 36 students were in computer science, 38 students were in blend, 23 students were in other, and eight students failed to disclose a major (unknown; not shown in Figure 1). Out of the blend students who applied to the program, 45% joined, making this applicant pool the most likely to join the program. This pool was followed by students from the other category (30%), the molecular life science category (24%), and the computer science category (17%). No students from the unknown category were asked to join the program. That blend applicants were most likely to join the program may have been both because they had a high probability of meeting the minimum course requirement and because they would be most eager to participate in a program directly related to their career path. The majors in the other category who joined the program were biostatistics, chemistry, chemical engineering, mathematics (2), and statistics. Four of the six students in this category had strong math backgrounds. One trend is noteworthy; the number of blend students who applied to the program increased each successive year. The data may reflect an increase in opportunities for students who want to pursue blend majors at their home campuses.
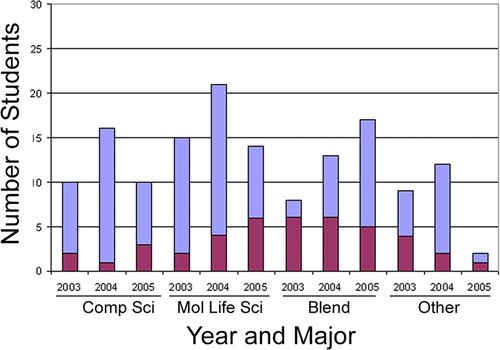
Figure 1. Majors who applied to and joined the SoCalBSI program. Blue bars, number of students who applied to the program; red bars, number of students who joined the program.
Of the 38 students accepted into the program, 40% were blend majors, 30% were molecular life science majors, 16% were other majors, and 14% were computer science majors. Overall, it seems that the blend students were either better qualified to join the program or were more willing to accept an offer from the program than students majoring in other subject areas. The number of molecular life science students who joined the program increased each successive year. This increase was true even though the number of molecular life science student applicants fell in the third year. This rising trend of molecular life science students joining the program may reflect an effort by these students to prepare themselves for a career in bioinformatics by successfully completing the necessary computer science course before applying.
Figure 2 shows the education levels of students who applied and joined the program. Students entering their junior year constituted the highest number of applicants (n = 50) and accounted for 34% of the total applicants with known education status (n = 145). Because these students must have completed a computer science and molecular biology course before completing their second year of college, the high percentage suggests that a high number of students are considering bioinformatics very early in their college careers. Students entering their first year of graduate school constituted the second highest number of applicants (n = 43), accounting for 30% of the total applicants. The percentages of undergraduate seniors and second-year graduates who applied to our program were 26 and 10%, respectively. Of the students who joined the program, the percentage of students at each of these education levels was as follows: junior, 37%; senior, 26%; first-year graduate, 30%; and second-year graduate, 7%. On the whole, the education level of students accepted into the program seemed related to the number of applicants from each education level, suggesting that students within each level were equally prepared for the program.
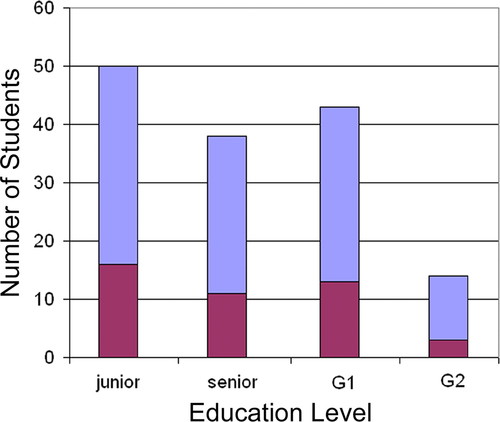
Figure 2. Education levels of students who applied to the program and of those who joined the program. Blue bars, number of students who applied to the program; red bars, number of students who joined the program. G1, first year of graduate school; G2, second year of graduate school.
A variety of factors seem to contribute to students' decisions to join SoCalBSI. To determine whether location influenced whether a student joined the program, we tallied the number of local and nonlocal residents (traveling >50 miles to our campus) that applied to SoCalBSI. Of the 147 applicants with known home campus locations, 63 were local and 84 were nonlocal. Although a greater number of students were nonlocal, a larger number of local students joined the program (21 local applicants joined the program and 17 nonlocal applicants joined the program). Furthermore, considering that five of the nonlocal students had relatives in southern California, suggesting that the students were familiar with the area, the number of “local” applicants who accepted increased even further. As expected for a large metropolitan area such as Los Angeles, the program seems to have had a greater success at attracting local applicants. That local students would not be required to pay for room and board also may have been a factor that led to higher numbers of local applicants joining the program. Of the 151 applicants for whom the application revealed gender, 89 were male and 62 were female. Of the 38 students who joined the program, 21 were male and 17 were female, suggesting a slight increase in the acceptance rate of females. Seven students who joined the program were underrepresented minorities, but it was unclear as to how many applicants were underrepresented minorities, because this question was not asked on the application.
Didactic Phase
The components of the didactic phase were chosen based on an article written by a pioneer in bioinformatics education, Russ Altman (Altman, 1998). The article lays out subject areas that should be covered in a graduate program in bioinformatics. Table 1 shows general subject areas, specialized subjects, the number of hours of the summer program devoted to each specialized subject area, and the instructor subject specialization. The total amount of instructor contact time with the students during the didactic phase was 80 h. In the general area of bioinformatics a large amount of time was spent on pairwise and multiple sequence alignment (8 h), because comparison of sequences has been the foundation on which many bioinformatics programs are built. Furthermore, extensive knowledge of sequence comparison programs helped the student understand the rationale for the programming project assigned during the didactic phase (see below). A substantial amount of time (6 h) was devoted to protein homology programs—specifically protein structure prediction programs. To understand this subject, students need to be familiar with the protein databank and with molecular structure-viewing programs. Students require time to become competent in manipulating the structures on the computer screen. Based on feedback from the students who attended our first summer program (see Program Assessment), we doubled the amount of time students spent on probability from 3 to 6 h, and we expanded the amount of time spent on microarrays from 4 to 9 h. All materials used in the didactic phase were placed online for the students and are freely available at http://instructional1.calstatela.edu/jmomand2/2005/curriculum/index.html. We found it particularly useful to include hyperlinks to commonly used databases and software programs on the SoCalBSI Web page.
| General area covered | Specific subject areas | No. of hours | Instructor specialization |
|---|---|---|---|
| Biology | Basic theoretical constructs in biology | 0.75 | Biochemistry |
| Molecular biology, genetics, cell biology | 0.75 | Biochemistry | |
| Computer science | Programming | 17.5 | Computer engineering |
| Math | Probability theory | 6 | Mathematics |
| Stochastic processes | 3 | Mathematics | |
| Optimization (e.g., expectation maximization) | 1 | Mathematics | |
| Specialized math and computer science subjects | Dynamic programming | 6 | Computer engineering |
| Ethics | Effects of technology on society | 1.5 | Philosophy |
| Privacy and security | 1.5 | Philosophy | |
| Bioinformatics | Pairwise sequence alignment | 5 | Biochemistry |
| Multiple sequence alignment | 3 | Biochemistry | |
| Phylogenetic trees | 3 | Biology | |
| Sequence feature extraction/annotation | 3 | Biochemistry | |
| Protein homology modeling | 6 | Biochemistry | |
| Protein threading | 1 | Biochemistry | |
| Integration of molecular biology databases | 3 | Biochemistry | |
| Microarray analysis | 9 | Biology | |
| Proteomics/signal transduction | 6 | Biochemistry | |
| Neural networks | 3 | Biochemistry | |
| Total | 80 |
We chose faculty at CSULA who could teach in those recommended subject areas. Two of the faculty, one a computer engineer and the other a biochemist, routinely teach the majority of these subject areas in a senior-level undergraduate bioinformatics course (4 quarter units) taught at CSULA. Additional subjects were taught by CSULA faculty with specialties in biology, mathematics, and philosophy. There were two 3-h didactic sessions per day, one session in the morning and one session in the afternoon. In general, the first 1.5 h of each session was used for a lecture presentation, and the next 1.5 h was devoted to a workshop that reinforced the lecture material.
An important component of the didactic phase curriculum was a computer programming project. Different components of a programming project were introduced early in the didactic phase, including data representations, data processing, file input/output, and simple user interfaces. The software engineering skills taught included algorithms, documentation, testing, and debugging. For the first year of the summer program, the C++ programming language was used. Based on feedback from research mentors (see Program Assessment), the Python programming language was used in subsequent years. Scripting languages such as Python and Perl are well suited to bioinformatics programming, because, compared with C++, scripting languages require fewer lines of code and are easier to write. Also, the scripting environments are more interactive and allow scientists to work more closely with the data (Dalke, 2004). Python was selected over Perl because it is a more formal language with real data structures and a large number of libraries, and it provides support for other high-level language extensions. In addition, Python is becoming a well-supported language in bioinformatics (Biopython, 2004).
Workshops associated with the programming project required students to develop simple bioinformatics programs such as a sliding window program to compute percentage of guanine and cytosine bases across a short sequence of DNA and a program to compute the PAM/BLOSUM score of single amino acid comparisons given a scoring matrix filename. Students also learned dynamic programming and recursion and implemented the longest common subsequence algorithm. These activities prepared students to complete a group-programming project. This project, in which students worked in pairs, required the students to implement a basic global and local sequence alignment tool and then add extensions for ends-free global alignment, affine gap penalties, and obtain an alignment from a series of sequences stored in simple database flat file. Wherever possible, a student more familiar with programming was paired with a student more familiar with molecular life science. The pairing of students with complementary strengths fosters the ability to communicate concepts between molecular life scientists and computer scientists—a hallmark of a competent bioinformaticist (Doom et al., 2003).
Part of the didactic phase was devoted to writing and presentation. Students made oral PowerPoint presentations of their programming projects to other students and the faculty at the end of the third week of the program. In addition to the programming project, students were given a choice between two writing assignments: one assignment was a summary of a recent research article in bioinformatics, and the other assignment was a review of a bioinformatics company that offers a software product. The articles were read and commented on by one of the instructors.
PROFESSIONAL DEVELOPMENT
An important objective of SoCalBSI was to develop skills that would help students obtain employment and sustain their careers. Friday afternoons of the first 9 weeks were devoted to these professional development activities. Bioinformaticists from industry and academia gave Friday seminars and had lunch with the students during which students were given the opportunity to ask about bioinformatics research, paths to careers, and current career opportunities. Later in the afternoons, students participated in activities that included the development of milestone charts to achieve research objectives, resume writing, and present PowerPoint presentations of their research projects.
Beginning on the Friday of the fourth week, students gave weekly progress reports on their research to peers and to some SoCalBSI faculty. These reports resulted in constructive exchanges of expertise between the students and also helped to identify the occasional mismatch of a student and research mentor. These mismatches were resolved early in the research phase and led to better experiences for the student and the mentor. In preparation for their final 15-min formal talks given at the end of the program, students gave practice presentations during the two prior Friday sessions that were critiqued by other students and two faculty with respect to the clarity of the explanation and presentation, quality and suitability of PowerPoint graphics, and speaking and presentation style. Formal critiques of the final presentations were made by faculty, students, and mentors. At their request, mentors were provided with the summary critiques of their students' presentations.
Research Phase
Students spent 7 weeks in a research internship at an academic institution or in industry. The students' final research presentations can be accessed at http://instructional1.calstatela.edu/jmomand2/2003/presentations/index.html, http://instructional1.calstatela.edu/jmomand2/2004/presentations/index.html, and http://instructional1.calstatela.edu/jmomand2/2005/presentations/index.html. With one exception, students worked at sites away from the CSULA campus in the greater Los Angeles area. To ensure that students and mentors would each receive the maximum benefit from the summer program, we developed a matching system based on expressed interest on the part of both research mentor and student. Before the start of the summer program, the completed applications of accepted students who were joining the program were made available to research mentors, who were asked to rank the students they felt would be a good fit for their research programs. Mentors were encouraged to rank students not only on level of experience but also on the students' stated interests. Research mentors were asked whether they would be interested in taking a pair of students, one student stronger in molecular biology skills and the other student stronger in computer science skills. Although most students were not paired, there were cases of very effective pairings as indicated by the quality of their research presentations. Students similarly ranked their choices for mentors. To aid students in making choices, mentors placed a description of their projects on the SoCalBSI website. The student-generated rankings of mentors and the mentor-generated rankings of students were used by the SoCalBSI faculty to determine the best combination of matches. Because the internships were offsite, a special consideration was transportation. Students who did not have cars were paired with students who did have cars and were going to the same institution. On rare occasions, students depended on public transportation to travel to their research sites.
PROGRAM ASSESSMENT
General Methodology
The purpose of assessment was to improve the quality of the program and to determine the subject areas in the didactic phase that required more background for students to perform well on their research projects. Assessment of the program involved both qualitative and quantitative techniques (Berg, 2001). To assess the quality of didactic instruction, a student opinion survey was administered at the end of the didactic phase. A survey/exit interview process was developed to improve the didactic phase curriculum in the areas of math, biology, and computer science. This process involved administration, to both students and mentors, of a formative survey at week 6 of the program followed by an exit interview or exit survey during the 10th week of the program. This design established two points for comparison regarding individual student achievement. Information from the first time point allowed individualized modification of the program for each student, whereas information from both time points helped to identify elements of the program that required improvement in order to better prepare professional bioinformaticists over the long term. Surveys and exit interviews were administered, and data were analyzed by an external investigator, one of us (B.K.), on this communication. The results were presented to the program principal investigators who used them to modify the SoCalBSI to attain its goal of training students for careers in bioinformatics.
Student Opinion Surveys
A forced answer survey of students' opinions of faculty teaching performances occurred on the final day of the didactic phase of the program (n = 14 students in 2003 and 2005 and n = 10 students in 2004). The questions and the results of this survey can be found at http://instructional1.calstatela.edu/jmomand2/3rd_Year/forms/assessment.htm. The summary data were used for program improvement, whereas the individual data were used by the instructors for teaching performance improvement. The statement with which the largest percentage of students most strongly agreed with was “The instructor interacted with the students in ways that were free of racial prejudice or discrimination” (82% in 2003, 84% in 2004, and 81% in 2005). In contrast, the statement with which the largest percentage of students either somewhat disagreed, disagreed with, or strongly disagreed was “The instructor clearly presented the lecture/workshop material” (2% in 2003, 9% in 2004, and 2% in 2005). In response to the question, “How would you rate this instructor's overall teaching ability?”, the percentage of students who rated instructors in the top two categories (excellent or very good) was 72% in 2003, 67% in 2004, and 68% in 2005. The data suggest that students were satisfied with the quality of instruction during the didactic phase of the program. One area that could be improved is the clarity of the presentations by the instructors during lectures and workshops. Based on the percentage of students who rated instructors in the top two categories for all statements in the survey, it seems that the quality of the program was high and consistent from 2003 through 2005.
Formative Surveys of Students for Self-Evaluation and of Research Mentors for Evaluation of Their Students
During the third week of the research phase of the program, both students and research mentors completed formative surveys consisting of a series of questions allowing open-ended responses. The questions on this survey were designed to elicit information that would 1) help identify characteristics of students and of the program curriculum critical for success in the research phase of the summer program, and 2) allow mentors and students to reflect on the work ethics of the students. The former information was used for improvement of the didactic phase of the summer program, student selection, and identification of skills and knowledge critical to success as a professional bioinformaticist, whereas the latter information was used for improvement of the professional development portion of the summer program. The formative self-evaluation survey answered by students, the formative student evaluation survey answered by research mentors, and summaries of answers to both surveys from 2003 to 2005 can be found at http://instructional1.calstatela.edu/jmomand2/index/assessment/index.html.
In total, 37 students responded to the survey between 2003 and 2005 (13 students responded in 2003, 10 students responded in 2004, and 14 students responded in 2005). In addition, 24 mentors responded to the survey between 2003 and 2005 (5 mentors responded in 2003, 5 mentors responded in 2004, and 14 mentors responded in 2005). Some mentors responded for multiple students. Figure 3 shows the top three student responses to formative self-evaluation questions regarding their own strengths in computer science, math, and biology that they bring to the research project. It also shows the top three research mentor responses to formative student evaluation survey questions regarding student strengths in these three areas. Specifically, research mentors were asked to comment on their students' preparedness for the project in computer knowledge, math knowledge, and biology knowledge. The response “other” indicates responses that could not be combined under a single heading. Figure 4 shows the top three student and instructor responses regarding students' weaknesses. Students were asked to answer the following question to identify weaknesses: What are your apparent weaknesses as they apply to the project with respect to: 1) computer knowledge? 2) math knowledge? 3) biology knowledge? Research mentors also were asked to answer the following questions to identify weakness of the student with regard to the research project: 1) What computer-related skills had you expected/hoped would be more fully developed in your student than they are? 2) What math concepts had you expected your student to have more completely internalized before their research experience? and 3) What biology concepts had you expected your student to have more completely internalized before their research experience?
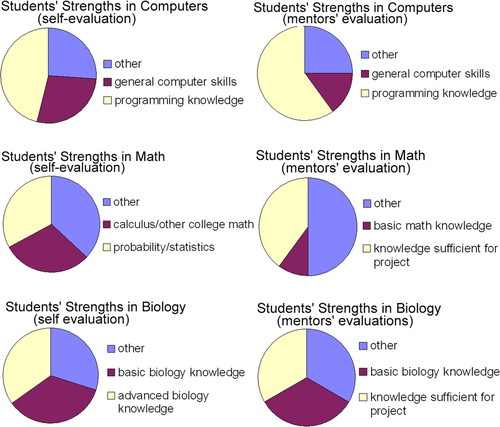
Figure 3. Comparison of results from student self-survey and research mentor survey of students in which students and research mentors were asked to assess the students' strengths in biology, math, and computer knowledge in preparation for the research project. Charts show top two responses for each skill category. The response “other” encompasses all responses that were individually less frequent than the top two responses. Self-assessment by students (n = 37); research mentor assessment of students (n = 30).
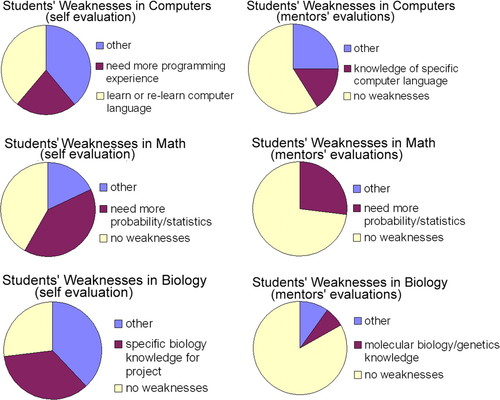
Figure 4. Comparison of student self-survey and research mentor survey of students where students and research mentors were asked to assess the students' weaknesses in biology, math, and computer knowledge with regard to preparation for the research project. Charts show top two responses for each skill category. The response “other” encompasses all responses that were individually less frequent than the top two responses. Self-assessment by students (n = 37); research mentor assessment of students (n = 30).
Students and research mentors both mentioned similar strengths and weaknesses in the formative survey, suggesting that the identified knowledge and skill set is essential to success as a professional bioinformaticist. The majority of students and research mentors noted that with regard to computer strengths, prior programming knowledge and general computer skills were essential for success on the research projects and that incomplete knowledge of a specific computer language could stall progress on a project. With regard to mathematics skills, the majority of research mentors felt that their students' math “knowledge was sufficient for the project,” and most students felt that their backgrounds in college-level math and/or probability and statistics prepared them adequately for their projects. Confirming this finding, the majority of both students and mentors indicated that students had no weaknesses with regard to mathematics skills/knowledge. However, when a weakness was identified, it was usually knowledge of probability and statistics. After the first summer, we addressed this weakness by increasing the math component in the didactic section. With regard to biology knowledge, mentors generally indicated that students' “knowledge was sufficient for the project” or that their “basic biology knowledge” was a strength, whereas students identified basic and advanced biology knowledge as strengths. Supporting this finding, the majority of mentors indicated that their students had no weaknesses with regard to biology. Many students agreed, but an equal proportion indicated that lack of “specific biology knowledge” could prevent progress on their project.
A set of questions posed in the formative survey centered around what knowledge was necessary for students to perform well on their assigned research projects. Students were asked “What necessary background was completely lacking or inadequate in your preparation for your internship?” The data showed that 30% of respondents (n = 37 from 2003 to 2005) indicated “nothing” (rank 1 in all 3 yr) and 19% of respondents indicated knowledge of “statistics and statistical packages” as a weakness (rank 2 in 2003, rank 3 in 2004, and tied for rank 3 in 2005). “From where should this knowledge be gained?” was a question asked of research mentors. Research mentors offered conflicting answers regarding whether skills and knowledge of probability and statistics should be acquired by students as part of their college curriculum, with 33% responding in the affirmative and 44% indicating that this information should be “acquired on the job.” Similarly, 33% of research mentor respondents indicated that specific knowledge of computer languages/skills should be acquired as part of the college curriculum, whereas 33% indicated that this knowledge could be “acquired on the job.”
Students cited similar prior course work or SoCalBSI workshops as essential to their success in their internships. Here, 26% of respondents (n = 37 from 2003 to 2005; rank 1 in 2003 and tied for rank 1 in 2004–2005) indicated that previous programming or software engineering courses were helpful in their internships, and 18% of the same respondents indicated that SoCalBSI didactic workshops provided such background.
Prior course work in molecular life science also was cited as of importance for research project progression. Here, 25% of respondents indicated that previous course work in molecular biology, biochemistry, or genetics was essential to success in the internship (rank 2 in 2003 and tied for rank 1 in 2004–2005), and 24% of the same respondents indicated that the SoCalBSI workshops on microarrays provided such background (rank 1 in all years). It should be noted that 30% of respondents felt that they did not “completely lack” anything necessary for success in their internships.
Student Exit Interviews
Individual student exit interviews of 20–30 min (n = 13 in 2003, n = 10 in 2004, and n = 14 in 2005) were conducted during the final week of SoCalBSI. The questions posed to the students were open ended and general. The questions were 1) What part of your involvement in the SoCalBSI met or exceeded your expectations? Please explain why or how.; 2) What part of your involvement in SoCalBSI disappointed you? Please explain your response and/or suggest ways to improve the program with respect to the issue that you have identified.; 3) Would you recommend to a colleague that they accept an internship in SoCalBSI? Please answer No, Yes, or Yes with reservations. Please explain your response.; and 4) Do you have any other comments about SoCalBSI that you would like to make at this time?
According to the student exit interview data, the didactic and research sections of the program and the overall program and its organization are strengths of the SoCalBSI program. This conclusion is supported by student exit survey data where 73% of respondents over 3 yr (n = 12 in 2003, n = 10 in 2004, and n = 14 in 2005; n = 37 students total) stated that SoCalBSI's didactic training is a strength of the program. Didactic training was the first-ranked item on the exit interview by students in 2003 and 2004 and the second-ranked item in 2005. The research phase of SoCalBSI also ranked high. In the interviews, 46% of respondents over 3 yr cited the research phase of SoCalBSI as exceeding expectations (n = 13 in 2003, n = 10 in 2004, and n = 14 in 2005; n = 37 total respondents,). This aspect of the program tied for second rank in 2003, was fourth in 2004, and was first in 2005. The “Overall program and its organization” was cited as an aspect of SoCalBSI that exceeded students' expectations from 2003 to 2005, with 43% of respondents indicating this as a strength of the program. It tied for second in 2003, tied for second in 2004, and was third in 2005.
OUTCOME OF SoCalBSI GRADUATES
The majority of students who completed the summer program are obtaining further education in a field related to bioinformatics. Of 38 graduates, nine graduates are currently in Ph.D. programs, 10 graduates are in master's programs, 11 graduates are in undergraduate programs, six graduates are in the workforce full time, and three graduates are involved in other activities. Out of the six graduates who are in the workforce full time, five graduates are working in bioinformatics or in an area where bioinformatics will be extensively used. One SoCalBSI graduate works full time outside the area of bioinformatics as a statistician. From the three students involved in other activities, one student is applying to medical school, one student is taking postbaccalaureate courses in preparation for pharmacy school, and one student is taking postbaccalaureate courses in psychology. In addition, four SoCalBSI graduates are working part time while pursuing their studies. Two of these graduates are working in the bioinformatics sector, and the other two graduates are working in the computer science sector.
Several of SoCalBSI graduates who are currently in formal graduate programs applied to these programs after completing the SoCalBSI program. Thus, it is feasible that a part of their career progression can be attributed to SoCalBSI. From the nine students in Ph.D. programs, six students began the application process after they attended SoCalBSI. Of the 10 SoCalBSI graduates currently in M.S. programs, four graduates applied to M.S. programs after completing the SoCalBSI program. Within the 38 SoCalBSI graduates, nine graduates are in the process of applying to graduate school. When SoCalBSI began, it was the stated goal that 75% of our students would enter a bioinformatics-based graduate program or a bioinformatics career. Although no former SoCalBSI students have yet completed their Ph.D. programs, from the graduate programs and careers chosen by SoCalBSI graduates, it is estimated that 34 of 38 SoCalBSI graduates (89%) are headed to a career that will either be directly related to the development of the bioinformatics field or in which bioinformatics will be a part of their job activities.
DISCUSSION
In this report, we have provided a detailed description of a 10-wk intensive summer program designed to prepare undergraduate and early graduate students for positions in the bioinformatics sector. Our program is unique because students who participate have the opportunity to perform research at one of nine research institutions from industry or academia. The feedback provided by students and their research mentors was used to ascertain a broad set of skills and knowledge that students required to succeed in bioinformatics research.
We also present an assessment strategy, including instruments (formative surveys found at our website, faculty evaluation and exit interview/focus group questions embedded in the text) that can be adapted for use in the evaluation of other bioinformatics programs. With the exception of generalized assessment strategies for individual courses (Centeno et al., 2003; Honts, 2003), to our knowledge no assessment strategies/instruments for a bioinformatics program with a didactic and research component have been published previously.
The results presented in this report suggest that our program could act as a model for other bioinformatics programs, particularly for campuses that do not have the resources to offer a bioinformatics degree or for those that want to address niche areas in the discipline (Zatz, 2002; Ranganathan, 2005). For example, the overall SoCalBSI program and its organization as well as the 3-wk didactic and 7-wk off-campus research phases were considered by students to be strengths of the program. With regard to didactic instruction, 67–98% of students responded positively (agreed or strongly agreed) with statements on student opinion surveys that asked questions regarding elements of teaching performance, 67–72% of students rated SoCalBSI instructors' overall teaching ability at the very good to excellent level, and the majority of students (73%) mentioned in the exit interview that the didactic phase was a strength of the program. Overall, it seems that the quality of didactic instruction has been high and consistent from the time the program started in 2003. In addition to the didactic phase, we report that both the research phase of SoCalBSI (46% of students noted it as a strength of the program) and the overall program and its organization (43% of students noted it as a strength) ranked high on student exit interviews.
Qualitative data generated from surveys and exit interviews/focus groups suggest a set of core skills and knowledge that are essential components to a bioinformatics curriculum (Figures 3 and 4). Identification of this skill and knowledge set is unique in the bioinformatics literature in that it involves an agreement between students and mentors regarding which skills and knowledge are essential to successful completion of a bioinformatics research project. Many scholars agree that the ability to accomplish such a research project is the key skill in bioinformatics (Brass, 2000; Pearson, 2001; Zatz, 2002; Counsell, 2003; Ranganathan, 2005), yet no prior study has established the extent to which students achieve this skill. Our data indicate that both mentors and students agree that the students' previous training coupled with the SoCalBSI didactic program adequately prepare students for bioinformatics research projects (Figures 3 and 4).
In our survey, both students and research mentors indicated that prior programming knowledge and general computer skills were essential to success on bioinformatics research projects; that general mathematics skills obtained through current undergraduate natural sciences programs were adequate for the project, although probability and statistics expertise could be improved; and that biology knowledge was adequate (although advanced or specific knowledge could be improved).
Student selection is an important aspect of any academic program and aids in the success of the program. The majority of students accepted into the SoCalBSI program were either equally balanced in molecular life science and computer science (e.g., cybernetics) or molecular life science majors (Figure 1), suggesting that intensive summer bioinformatics programs might concentrate future recruitment efforts on these students. In contrast (with the exception of second-year graduate students), the number of individuals who joined the program were approximately evenly distributed across education level (junior undergraduate through first-year graduate; Figure 2), suggesting that no specific educational level within this group should be targeted for recruitment by summer bioinformatics programs. Other factors may powerfully impact the make-up of the student population of the SoCalBSI. Local students were more likely to join the program than were nonlocal students, whereas males and females seemed approximately equally likely to join. It is unclear what impact minority status had on the likelihood that a student would join the program.
SoCalBSI graduates seem to be motivated to continue education in a bioinformatics-related field; we estimate that 34 of 38 graduates (89%) are in a career trajectory that will use bioinformatics. A majority of these students are either currently in graduate programs or are planning to attend graduate school in a bioinformatics-related field. Another National Science Foundation–National Institutes of Health-sponsored Bioengineering and Bioinformatics Summer Institute located at the University of Pittsburgh (Pittsburgh, PA) recently published an account of their program that, similar to SoCalBSI, consists of a two-phase training program with a focus in computational biology (Munshi et al., 2006). According to a survey of its student participants, 62% indicated that they would either definitely or possibly/likely enter the field of computational biology. It is difficult to conclude that SoCalBSI or the program at the University of Pittsburgh have heightened an interest in bioinformatics. Students recruited to both programs were likely to be highly motivated to pursue this field a priori.
Our results showing that a high percentage of students who plan to continue their education/careers in a bioinformatics-related field are not surprising. A survey of 1088 undergraduates with summer research experiences found that such experiences either confirmed or did not alter plans for postgraduate education in at least 87% of the cases (Lopatto, 2004). This suggests that summer programs with strong research components can affirm students' career paths. It will be important to continue to track students from SoCalBSI or the program at Pittsburgh to determine the percentages that actually enter into the bioinformatics or the computational biology workforce.
ACCESSING MATERIALS
The specific link for the curriculum is http://instructional1.calstatela.edu/jmomand2/2005/curriculum/index.html.
Student Perception of Faculty Performance Surveys were constructed at http://nss-nemo.calstatela.edu/ad/Assessment.
Complete results of formative surveys and assessment outcomes can be found at http://instructional1.calstatela.edu/jmomand2/index/assessment/index.html.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation–National Institutes of Health Grant EEC-0234129 and the Los Angeles/Orange County Biotechnology Center.



