A Western Blot-based Investigation of the Yeast Secretory Pathway Designed for an Intermediate-Level Undergraduate Cell Biology Laboratory
Abstract
The movement of newly synthesized proteins through the endomembrane system of eukaryotic cells, often referred to generally as the secretory pathway, is a topic covered in most intermediate-level undergraduate cell biology courses. An article previously published in this journal described a laboratory exercise in which yeast mutants defective in two distinct steps of protein secretion were differentiated using a genetic reporter designed specifically to identify defects in the first step of the pathway, the insertion of proteins into the endoplasmic reticulum (Vallen, 2002). We have developed two versions of a Western blotting assay that serves as a second way of distinguishing the two secretory mutants, which we pair with the genetic assay in a 3-wk laboratory module. A quiz administered before and after students participated in the lab activities revealed significant postlab gains in their understanding of the secretory pathway and experimental techniques used to study it. A second survey administered at the end of the lab module assessed student perceptions of the efficacy of the lab activities; the results of this survey indicated that the experiments were successful in meeting a set of educational goals defined by the instructor.
INTRODUCTION
Studying the Yeast Secretory Pathway in an Undergraduate Laboratory Course
The eukaryotic secretory pathway comprises the events by which proteins are inserted into the endoplasmic reticulum (ER), trafficked between the various membrane-bound organelles of the endomembrane system, and brought to the cell surface. This fundamental cellular pathway is a topic taught in most introductory and intermediate college cell biology courses. Our current understanding of protein secretion is based upon the combined knowledge gained from discoveries made using traditional cytological methods, more modern imaging techniques, in vitro reconstitution experiments, and genetic studies (Karp, 2005). Consequently, the secretory pathway functions particularly well as a means for introducing students to the various experimental approaches that can be used to learn about cellular processes.
Genetic studies of the eukaryotic secretory pathway have primarily used the model organism Saccharomyces cerevisiae, or budding yeast. Yeast is an ideal model system for undergraduate laboratories because it can be easily manipulated in genetic, biochemical, and cytological experiments; it grows rapidly under inexpensive conditions; and its sequenced genome is highly annotated. An article describing a laboratory series using a genetic reporter system to study the secretory pathway in yeast was published previously in this journal (Vallen, 2002). The current article describes a Western blotting experiment that has been developed for use in conjunction with the previously described genetic assay as a second means of investigating the particular steps of the secretory pathway that are blocked in two temperature-sensitive mutant yeast strains (sec mutants).
The two mutant strains that are used in the assays are sec61-1 and sec18-1. The sec61-1 strain bears a mutation in a gene that encodes a component of the ER translocation machinery, Sec61p, and it is therefore defective for the first step in the secretory pathway (Deshaies et al., 1991). The sec18-1 strain, in contrast, is defective in vesicle transport from the ER to the Golgi complex due to a mutation in the gene that encodes the yeast homolog of the mammalian NEM-sensitive factor Sec18p, which is an ATPase required for vesicle fusion (Wilson et al., 1989; Peters et al., 1990).
As described by Vallen (2002), the genetic assay uses a reporter gene (ss-HIS4, a gene that encodes a histidine biosynthetic enzyme fused to an ER signal sequence) that was first used by Deshaies and Schekman (1987) in a screen designed to identify genes that functioned in the cotranslational insertion of proteins into the ER. SEC61 was one gene discovered in this screen. Because of the specific nature of the reporter gene, the genetic assay allows students to obtain corroborating evidence that the sec61-1 strain has a defect in ER insertion; however, they do not gain any insight to the precise nature of the sec18-1 defect beyond the fact that it does not affect ER insertion. The Western blotting experiments described here complement the genetic assay by providing more direct information about the steps that are blocked in both sec mutants.
Following Protein Glycosylation via Western Blotting
As proteins move through the secretory pathway, they are modified in various ways, including by glycosylation, which is the covalent attachment of sugar chains to amino acids contained within specific target sequences. The first, or core, glycosylation events occur in the ER, where branched sugar chains are attached en bloc to asparagine residues that fall within the amino acid sequence Asn-X-Ser/Thr (Karp, 2005). This type of glycosylation is referred to as N-linked (N for asparagine). As proteins move from the ER through the various compartments of the Golgi complex, the core N-linked sugar chains undergo modifications, including the trimming of some of the terminal sugar moieties and, for many proteins, the addition of other sugar monomers to the chains. Some proteins also undergo a second type of sugar modification in the Golgi complex, O-linked glycosylation, which modifies the hydroxyl groups of serine or threonine residues (Karp, 2005). The enzymes that carry out these modifications are localized to specific compartments of the endomembrane system; therefore, changes in a protein's molecular weight due to glycosylation and other processing events serve as indicators of the protein's progress through the secretory pathway (Esmon et al., 1981). Consequently, a Western blot using an antibody that recognizes a particular secreted or cell-surface protein can be used to determine which form of the protein accumulates in a given sec mutant; this information can in turn reveal the intracellular compartment in which the protein is trapped and therefore the specific secretory step that is compromised in the mutant.
This article describes Western blot experiments that detect two different proteins, pre-pro-α-factor (pp-α-F) and invertase, as a means for obtaining corroborating evidence of the specific secretory steps that are defective in the sec18-1 and sec61-1 mutant yeast strains. Both of these proteins have been used extensively as model secreted proteins (Esmon et al., 1981; Julius et al., 1984). pp-α-F is the precursor to the mating pheromone produced by haploid yeast of the MATα mating type. Mature, secreted α-factor is a polypeptide of only 13 amino acids, but it is synthesized as a larger precursor protein that first acquires N-linked glycosylation in the ER, and then it is processed to its mature form in proteolytic steps that occur in the Golgi and in secretory vesicles (Julius et al., 1984). Invertase is an enzyme encoded by the yeast SUC2 gene that hydrolyzes the disaccharide sucrose to its monosaccharide components, glucose and fructose, and is therefore required for yeast to grow on sucrose-containing media. Yeast express two types of invertase: a cytoplasmic form that is produced constitutively and a cell surface form that is repressed when cells are growing in glucose-containing medium but is strongly up-regulated when cells are shifted to low-glucose medium (Dodyk and Rothstein, 1964). These two forms of invertase are produced from two different mRNAs transcribed from the SUC2 gene, one form that encodes an N-terminal ER signal sequence and one form that lacks this sequence (Perlman and Halvorson, 1981; Carlson and Botstein, 1982). The experiment described here uses a modified SUC2 gene, SUC2–13myc, which causes both forms of the protein to be expressed with a C-terminal epitope tag that can be recognized by a commercially available monoclonal antibody.
The Course Context
A 3-wk laboratory module combining the ss-HIS4 reporter assay and one of the two versions of the Western blot has been implemented four times in the course Biological Sciences 220 (BISC 220, Cellular Physiology) at Wellesley College during the period from spring 2004 to spring 2007, involving approximately 150 students in total. Wellesley College is a women's liberal arts college with an overall population of approximately 2300 undergraduates; ∼80 students each year graduate with majors in areas of the life sciences. BISC 220 is an intermediate-level cell biology course that begins with an introduction to protein biochemistry, including protein structure and enzymology, and then covers the molecular basis for major cellular processes such as intracellular protein transport, cell signaling, cytoskeletal dynamics, and the cell division cycle. Most of the students are sophomores or juniors. The course is required for biological chemistry majors and is also taken by many biological sciences and neuroscience majors. Introductory-level cell biology and two units of college chemistry are prerequisites, but a significant number of students have also taken an intermediate-level genetics course that combines classical and molecular genetics. The associated laboratory is scheduled for 3.5 h/wk; each lab section has a maximum of 12 students and is taught either by one of the lecturing faculty or a master's level laboratory instructor.
The yeast secretory pathway module follows a 4-wk series in which students purify an epitope-tagged enzyme and perform kinetic studies on its catalytic activity. The students assess their purification using Coomassie-stained gels, so they have already learned the theory behind SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and how to calculate the estimated molecular weight of a protein based on its migration distance. The students have also been exposed to the Western blotting technique in lecture, but many have not had prior hands-on experience with it.
Evaluation Methods
When the experimental series described here was added to the BISC 220 lab curriculum, the instructors hoped that it would facilitate three main gains in the students who participated in it: 1) an improved understanding of the processes by which proteins are secreted from the cell or targeted to the various compartments of the endomembrane system; 2) familiarity with the ways that yeast genetics can be used to understand fundamental problems in cell biology; and 3) experience with the important experimental technique of Western blotting and an understanding of how it can be used to study various modified forms of a protein. Although the lab had been used three times previously with perceived success from the instructors' viewpoints, its efficacy in meeting these educational goals was not formally assessed until the spring 2007 semester. In that iteration of the lab module, we implemented two evaluation tools: a knowledge survey administered before and after the module and a student attitude survey administered at the end of module. The former tool was designed to measure the effect of the lab activities on the students' understanding of secretory pathway function and experimental techniques used in the lab, and the latter tool assessed the students' perceptions of the extent to which the lab activities facilitated their learning. The results of the surveys provide evidence of the educational efficacy of the lab series.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
The yeast strains used in this study are listed in Table 1; all are available upon request. LY527, LY689, and LY651 were kindly provided by Elizabeth Vallen (Swarthmore College); JHY452 was a gift from Vlad Denic and Jonathan Weissman (University of California, San Francisco). The HOL1–1 allele carried by LY527, LY689, and LY651 is required for entry of histidinol into cells (Gaber et al., 1990), which is important for the ss-HIS4 genetic reporter assay (Vallen, 2002); because these three strains are all MATα, they express the pp-α-F detected in one version of the Western blot experiment. JHY453, JHY454, and JHY455 contain the SUC2–13myc gene, which encodes the myc-tagged invertase (Suc2p-13myc) detected in the other version of the Western blot experiment. Standard yeast culture methods and growth media were used (Sherman, 1991); specific details related to the yeast growth assay media have been described previously (Vallen, 2002). We purchased the synthetic amino acid dropout mixtures required for the genetic reporter assay from United States Biological (Swampscott, MA; http://www.usbio.net), which we have found to be more reliable than other suppliers. For the pp-α-F experiment, cells were grown at 25°C to exponential phase (OD600 approximately 1.0) in yeast extract peptone dextrose (YPD) medium, and then either shifted to 37°C or maintained at 25°C for 1 h before harvesting. For the invertase experiment, cells were grown at 25°C to exponential phase (OD600 approximately 1.0) in standard YPD medium (containing 2% dextrose), then pelleted and resuspended in low-dextrose YPD (0.1% dextrose) to induce expression of the secreted form of invertase; the low-dextrose cultures were either shifted to 37°C or maintained at 25°C for 3 h before harvesting.
| Strain name | Partial genotypea | Reference/source |
|---|---|---|
| LY527 (WT) | MATα ura3 his4 leu2 trp1 HOL1-1 | Vallen, 2002 |
| LY689 | MATα sec18-1 ura3 his4 HOL1-1 | Vallen, 2002 |
| LY651 | MATα sec61-1 ura3 leu2 trp1 his4 HOL1-1 | Vallen, 2002 |
| JHY452 | MATa SUC2-13myc:HIS3 ura3 leu2 trp1 ade2 his3 | J. Weissman, University of California, San Francisco |
| JHY453 | MATa SUC2-13myc:HIS3 ura3 leu2 trp1 | This study |
| JHY454 | MATα SUC2-13myc:HIS3 sec18-1 ura3 | This study |
| JHY455 | MATa SUC2-13myc:HIS3 sec61-1 ura3 leu2 trp1 | This study |
| JHY359 | MAT a ura3 leu2 trp1 his3 | J. Hood-DeGrenier, unpublished |
Cell Lysate Preparation and SDS-PAGE
Cell pellets derived from 10 ml of culture were lysed in SDS sample buffer (1% SDS, 2.5% glycerol, 25 mM Tris-HCl, pH 6.8, 50 mM dithiothreitol, and 0.05% bromphenol blue) using a FastPrep cell lysis instrument (MP Biomedicals, formerly Qbiogene, Solon, OH; http://www.mpbio.com) as described in the Protocols section of Supplemental Material A1. The glass beads used in the lysis were from Sigma-Aldrich (St. Louis, MO; http://www.sigmaaldrich.com, catalog no. G-8772). Substituting 5 min of vortexing at top speed for the FastPrep lysis resulted in lower-quality lysates, but another type of bead beater should work equally well. We also tried lysing the cells in a buffer containing a milder detergent with protease inhibitors rather than SDS sample buffer and achieved similar results (protease inhibitors were not used when the cells were lysed in SDS sample buffer). For the pp-α-F experiment, the lysates were separated on 12.5% Tris-HCl SDS-polyacrylamide gels as described in Supplemental Material A1; 7.5% Tris-HCl SDS-polyacrylamide gels were used for the invertase experiment (see Supplemental Material A2). (Note: you may need to adjust the volume of lysis buffer and/or the volume loaded on the gel to optimize the quality of your results.) Precast gels were purchased from Bio-Rad (Hercules, CA; http://www.Bio-Rad.com); the Western blots shown in Figure 1 were performed using Bio-Rad Criterion gels, but the students used Bio-Rad mini-gels and achieved similar results. Precision Plus Dual Color Prestained Protein Standards (Bio-Rad, catalog no. 161-0374) were used in both experiments.
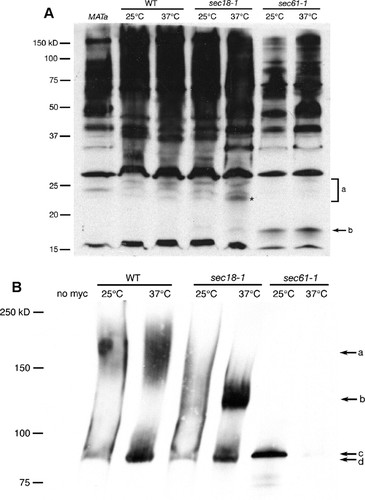
Figure 1. (A) Western blot detecting pp-α-F in lysates from WT (LY527), sec18-1 (LY689), and sec61-1 (LY651) yeast strains grown continuously at 25°C or shifted to 37°C for 1 h. “MATa” lane contains a lysate from cells that do not produce pp-α-F (JHY359) and thus serves as a negative control for antibody specificity. Bands contained in the a bracket represent glycosylated forms of pp-α-F that exist in the ER, with the asterisk indicating the singly glycosylated pp-α-F; b points to unglycosylated pp-α-F. Relative mobilities of molecular-weight-standard bands are shown on the left. (B) Western blot detecting 13xmyc-tagged invertase in lysates from SUC2–13myc (WT; JHY453), sec18-1 SUC2–13myc (JHY454), and sec61-1 SUC2–13myc (JHY455) yeast strains transferred from regular YPD to low-glucose YPD and grown for 3 h at either 25 or 37°C. “No myc” lane contains a lysate from cells with an untagged SUC2 allele (JHY359) and thus serves as a negative control for antibody specificity. Arrow a indicates the mature form of invertase; b indicates the ER-modified form; c indicates the unmodified, secreted form; and d indicates the cytosolic form of invertase that is produced constitutively.
Western Blotting
Separated proteins were transferred to Hybond-ECL nitrocellulose membrane (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom, formerly Amersham Biosciences; http://www.gehealthcare.com, catalog no. RPN303D) by using a Bio-Rad protein transfer apparatus as described in Supplemental Material A1. The transfer buffer contained 12.5 mM Tris base and 96 mM glycine. After the transfer, the nitrocellulose was stained with Ponceau S (Sigma-Aldrich, catalog no. P-7170) for approximately 10 s, and then the membrane was rinsed with distilled water to allow comparison of the relative amounts of protein loaded in each lane and to check for any regions of the gels that did not transfer efficiently. The membranes were wrapped in plastic wrap and stored at 4°C for 2 wk before immunodetection, which was performed as described in Supplemental Material B. The blocking buffer contained 5% dry milk powder in 1X phosphate-buffered saline (140 mM sodium chloride, 2.7 mM potassium chloride, 10 mM dibasic sodium phosphate, and 2 mM monobasic potassium phosphate) plus 0.25% Tween. The primary antibody used to detect pp-α-F was a protein A affinity-purified rabbit polyclonal antibody raised against a peptide corresponding to amino acids 112–125 of the MFα1gene product that was prepared by GenScript (Piscataway, NJ; http://genscript.com). Limited quantities of this antibody will be available upon request. Suc2p-13myc was detected using a mouse monoclonal anti-c-myc 9E10 antibody (Sigma-Aldrich, catalog no. M-4439). (Note: you may want to try several different antibody dilutions to obtain optimal results.) Horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) (catalog no. 111-035-003) and goat anti-mouse IgG (catalog no. 115-035-062) were from Jackson ImmunoResearch Laboratories, (West Grove, PA; http://www.jacksonimmuno.com) and were stored in 50% glycerol after reconstitution as recommended by the manufacturer (dilution factors specified in Supplemental Material B refer to the 50% glycerol stocks). HRP was detected by enhanced chemiluminescence using the ECL Western Blotting Analysis System from GE Healthcare (catalog no. RPN2109) and Kodak XAR-5 film (ordered from Sigma-Aldrich), which was developed using an AutoTank automatic x-ray film processor from Fischer Industries (Geneva, IL; http://www.fischerind.com). GLOGOS II glow-in-the-dark tape (Stratagene, La Jolla, CA, catalog no. 420201; http://www.stratagene.com) was used to facilitate proper alignment of the developed film with the nitrocellulose for the purpose of determining molecular weight marker position. (Note: you will need to optimize the film exposure times; in different trials of the experiment the optimal times were as short as 15 s or as long as 10 min, depending on the amount of protein loaded and the antibody dilution.)
The Laboratory Schedule
Typically, the students have not seen the secretory pathway in lecture by the start of the lab series (but it is covered in lecture by the end of the 3-wk module); therefore, we begin with a brief overview of the pathway as an introduction to the series. This introduction includes a discussion of the concept of temperature-sensitive mutants and the rationale behind the ss-HIS4 reporter construct. In our version of the ss-HIS4 genetic reporter assay, the students are given the required transformed yeast strains rather than doing the plasmid transformations themselves; they spot the transformed strains onto the appropriate growth media during week 1 of the module, and they score their results in week 2. For this assay, the students are told which yeast strain is which, but they are required to discover the functions of the Sec18p and Sec61p proteins themselves by searching the Saccharomyces Genome Database (http://www.yeastgenome.org). They are also asked to perform a BLAST search of the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) to identify the closest human homologs of the Sec18p and Sec61p proteins. The students do these exercises in pairs during the first lab period, and then they predict the results of the genetic assay as an individual homework assignment due the following week. The students record their growth predictions in a table like that described by Vallen (2002). In addition to predicting whether each strain will grow under each condition, they are asked to explain their predictions by indicating whether a lack of growth can be attributed to temperature sensitivity of the strain, to the yeast not expressing an enzyme required to synthesize an essential compound that is missing from the medium, or to mislocalization of such an enzyme. A shorthand code is used for these explanations (see Supplemental Material A). The students are directed to the article by Deshaies and Schekman (1987) that first described the reporter system as a reference to assist their understanding of the experimental scheme.
In week 2, the students prepare the yeast lysates, perform the SDS-PAGE, and do the Western blot transfer. The yeast pellets are prepared in advance and frozen at −20°C in individual screw-cap microcentrifuge tubes so that they are ready at the start of the lab. Preparing the lysates takes approximately 30 min, loading and running the gels takes 45–60 min, and setting up the Western blot transfers takes another 30 min. While the gels are running, the students evaluate their genetic results with some assistance from the instructor; they are asked to score the actual results without looking at their predictions and then to compare the two and try to explain any discrepancies. The immunodetection portion of the Western blot is performed in week 3. The course lab manual sections for weeks 2 and 3 are included in Supplemental Materials B1, B2, and C (with B1 and B2 being specific for the pp-α-F and invertase versions of the Western blot, respectively); the lab manual section for week 1 was omitted because the yeast growth assay was performed essentially as described by Vallen (2002). Instructions for the computer exercises used in week 1 are included in Supplemental Material D.
At the end of the 3-wk lab series, the students are required to write a full lab report in the style of a scientific paper that includes their data from both the genetic reporter assay and the Western blot. This is the second such report required during the semester. Throughout the experimental part of the lab module, the students work in pairs, but the lab report is written independently. Because the students start out knowing the functions of the SEC18 and SEC61 genes before they obtain their own results, they are told to write from the perspective of using the two assays to confirm what has already been published about the genes, rather than artificially describing their results as novel findings.
Laboratory Costs
Many of the reagents and materials required for the Western blotting experiment are rather expensive, but some reagents are purchased in quantities that are sufficient for more than one course-worth of students, and some alternatives exist that can reduce costs significantly. We estimate that the cost per student (for a class of 36) for the Western blot in spring 2007 was $26, excluding the cost of generating the anti-pp-α-F antibody. The anti-myc antibody used for the Suc2p-13myc blot costs $240 for 100 μl. Used at a 1:7500 dilution, this quantity is sufficient for approximately 80 blots; if necessary, the diluted antibody can be saved and reused for multiple blots for a period of 1 to 2 wk, and the undiluted antibody can be frozen in small aliquots at −20°C for at least 2 yr. The HRP-conjugated secondary antibodies cost $79 for 2 ml, which will last a very long time when stored as a 50% glycerol stock at −20 or −80°C. Apart from the antibodies, the most expensive items for the Western blot lab as we execute it are the ECL detection reagent ($223), the molecular weight marker ($120), and the prepoured gels ($10 each). Although we have not tried this method, alkaline phosphatase Western detection reagents would be cheaper; furthermore, more inexpensive molecular weight markers are also available, and costs could be cut by pouring your own gels. Therefore, after the first execution of this lab, the costs will be lower, and alternatives exist to make it feasible for instructors working with different budget constraints. The yeast growth assay portion of the lab series is relatively inexpensive, with media reagents costing approximately $300 for amounts that last us 5 yr.
Evaluation of Student Outcomes
Formal evaluation of this lab series was conducted in the spring 2007 semester with a population of 34 students enrolled in three lab sections. The pp-α-F version of the Western blot was used in that semester. The two surveys used in the assessment are included in Supplemental Materials E and F. The Knowledge and Understanding (KO) survey (which was administered both at the beginning and the end of the module) consisted of five multiple-choice questions with four choices each, the last of which was always “I have no idea,” and three true/false questions that also included the “I have no idea” option. The “no idea” choice was included in an effort to obtain an accurate picture of the students' knowledge that was not obscured by possible artifacts caused by random guessing, and the instructions to the students clearly stated that they should select this response rather than guessing. The Perceived Efficacy (PE) survey (administered at the end of the module) asked students to use a 5-point Likert scale (Strongly Agree, Agree, Neither Agree Nor Disagree, Disagree, or Strongly Disagree; Likert, 1932) to rate the degree to which the lab module helped them achieve certain learning goals. The number of respondents to the KO pre- and postsurveys and the PE survey were 27, 28, and 26, respectively. Participation in the surveys was voluntary, anonymous, and did not involve any personal information, so the study was exempt from review by the college's Institutional Review Board.
RESULTS
Expected Western Blot Results
Loss of Sec61p function causes normally secreted proteins to be mislocalized to the cytosol, rather than being directed through the secretory pathway, whereas loss of Sec18p function causes normally secreted proteins to be trapped in the ER. Both model secreted substrates, pp-α-F and invertase, undergo cotranslational N-linked glycosylation in the ER; thus, the unmodified, full translation products of the MFα1 and SUC2–13myc genes should not be present in wild-type (WT) cells. One would predict that loss of Sec61p function would lead to accumulation of the unmodified, translated forms of these proteins and that these unmodified proteins would exhibit greater mobility in an SDS-PAGE separation than the ER forms that can be detected in WT cells. Loss of Sec18p function, on the other hand, would be expected to cause greater accumulation of the ER forms of the two proteins than is seen in WT cells. ER-trapped pp-α-F would have a higher molecular weight than other forms of the protein because pp-α-F glycosylation occurs only in the ER, and the protein undergoes proteolytic processing in the Golgi and later compartments. In contrast, ER-trapped invertase would have an intermediate molecular weight compared with the other forms of the protein, because invertase acquires carbohydrate modifications in both the ER and the Golgi. The mature forms of α-factor and invertase would be expected to be present only in cells in which both Sec61p and Sec18p are functioning at least partially. The mature α-factor is only 3.4 kDa, so it would be expected to run off the 12.5% polyacrylamide gels used in the pp-α-F experiment and therefore not to be visualized; the mature invertase, in contrast, would be expected to be the predominant form of invertase in WT cells and to have the lowest mobility of any of the forms of invertase detected by Western blotting.
pp-α-F Western Blot
Figure 1A shows an example of an anti-pp-α-F Western blot analyzing samples from WT, sec18-1, and sec61-1 cells grown continuously at 25°C or shifted to 37°C for 1 h. A band that migrates between the 15- and 20-kDa molecular weight markers (Figure 1A, arrow b) is present in both sec61-1 lanes, but not in the WT or sec18-1 lanes. The size of this band is consistent with the predicted molecular weight of the unmodified pp-α-F polypeptide, which is 18.6-kDa (Julius et al., 1984). A “ladder” of three bands in the molecular weight range of 20–26 kDa (Figure 1A, bracket a) is visible in all of the WT and sec18-1 lanes, but these bands are more pronounced in the sec18-1 37°C sample; in particular, the two bands with lower molecular weights in this set are significantly more abundant in the sec18-1 37°C lane than either of the WT lanes or the sec18-1 25°C lane. The sizes of these bands correlate with the predicted sizes of the ER form of pp-α-F modified with one, two, or three core N-linked oligosaccharide chains (Julius et al., 1984). These forms are almost totally absent from the sec61-1 samples, which is consistent with the inability of pp-α-F to reach the ER lumen in this strain. The increased abundance of these bands in the sec18-1 37°C sample as compared with WT and the sec18-1 25°C cells reflects the inability of proteins to move out of the ER to later compartments of the secretory pathway when Sec18 function is compromised.
Invertase Western Blot
Figure 1B shows an example of an anti-myc Western blot analyzing samples from WT SUC2-13myc, sec18-1 SUC2-13myc, and sec61-1 SUC2-13myc cells grown continuously at 25°C or shifted to 37°C for 3 h. There are two forms of Suc2p-13myc present in the WT cells: one band located between the 75- and 100-kDa molecular weight markers (Figure 1Bd), which presumably corresponds to the cytosolic form of invertase, and a second form that migrates in the 140- to 200-kDa range (Figure 1Ba). Although we do not know the exact sequence of the 13myc tag, including spacer sequences, we predict that it should add 20–25 kDa to the molecular weight of invertase; thus, the observed sizes for the cytosolic and mature forms of Suc2p-13myc are consistent with the known sizes of these forms of the untagged protein, which are 61- and 120- to 175-kDa, respectively (Novick et al., 1981; Ferro-Novick et al., 1984; Deshaies and Schekman, 1987). The sec18-1 37°C sample shows a large accumulation of an intermediate molecular weight form of the protein (Figure 1Bb), the size of which is consistent with it being the core N-linked glycosylated ER form of the protein (approximately 80 kDa for untagged invertase). As for pp-α-F, there is a band in the sec61-1 25°C lane that is absent from the other lanes (Figure 1Bc). This band is slightly higher than band d seen in the samples from the WT and sec18-1 strains, which is consistent with “d” being the constitutively expressed cytoplasmic form of invertase that lacks the ER signal sequence and “c” being the secreted form with the signal sequence that is trapped in the cytosol in sec61-1 cells.
Evaluation of Student Outcomes
The success of the 3-wk lab series in achieving the educational goals outlined in the Introduction was assessed by administering pre- and postlab KO surveys and a postlab PE survey to students in the most recent BISC 220 class (see Materials and Methods and Supplemental Materials E and F). Significantly more students were able to identify the correct answer postlab compared with prelab for the majority of the questions on the KO survey (Figure 2). This was particularly true for the questions that addressed the function of signal sequences, the effect of glycosylation on a protein's molecular weight, the advantages of conditional mutations, the compartments in which glycosylation occurs, and the analysis of glycosylated proteins by Western blotting (Supplemental Material E, questions 1–3, 7, and 8). Because the basics of Western blotting had already been discussed in lecture before the start of the lab module, nearly 80% of students already understood the function of the secondary antibody in a Western blot procedure at the start of the lab (question 5), but a postlab gain was also seen on that question. In the prelab survey, <50% of students chose the correct answer for four of the eight questions (3, 6, 7, and 8); this number was reduced to one in the postlab survey (question 6), and even for that question, the percentage of correct responses on the postsurvey was more than double that on the presurvey. Question 4, which assessed students' basic understanding of plasmids and their experimental use, was the only question that showed a very minimal postsurvey gain in correct answers. This was surprising, because plasmids were an integral part of the genetic reporter assay, but it may have been due to students misreading the question (see Discussion).
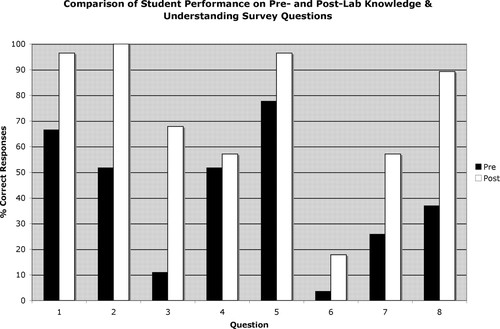
Figure 2. Comparison of the percentages of students who answered each of the nine questions on the knowledge survey correctly at the beginning of lab module (Pre) and after its completion (Post).
In addition to the significant gains in correct answers on the post- versus the prelab survey, there was an even more substantial drop in the percentages of students who responded “I have no idea” to any of the questions on the postsurvey (Figure 3). In the presurvey, >30% of students felt uncertain enough to choose this answer for four of the eight questions; in the postsurvey, no question generated significant uncertainty for more than two students in the entire survey population (7%). Interestingly, for the question on which the students performed the worst on the postsurvey (question 6), only 7% recognized that they did not know the correct answer, whereas 76% chose the incorrect answer with some confidence.
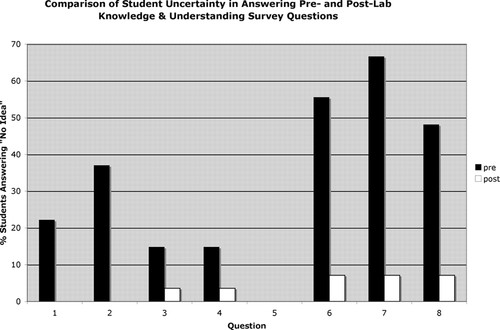
Figure 3. Comparison of the percentages of students who responded “I have no idea” to each of the nine questions on the knowledge survey at the beginning of lab module (pre) and after its completion (post).
The PE survey (see Supplemental Material F) asked students to use a 5-point Likert scale (Likert, 1932) to rate the extent to which they agreed that particular aspects of the lab module met their desired instructional goals (Figure 4). For all eight questions in this survey, 69% or more of students “Agreed” or “Strongly Agreed” that the educational goal was achieved; for five of the questions (1, 2, 6, 7, and 8), this number was >84%. In addition, no more than one student “Strongly Disagreed” that any of the individual educational goals was achieved, and only on one question (2) did more than one student choose either “Disagree” or “Strongly Disagree.” Most notably, students overwhelmingly agreed that the lab improved their understanding of the secretory pathway and that they would not have understood the two assays used to investigate the secretory pathway as well if they had just listened to lectures about them rather than performing the assays themselves (see responses to questions 1 and 3). There was also a very strong consensus among the students that making formal predictions about the outcomes of the two assays before seeing the actual results and completing the lab series with a written lab report augmented their learning (see responses to questions 7 and 8). In addition, students felt that the Western blot performed in the lab helped them to better understand the technique in general and how it might be used in other experimental scenarios (see responses to questions 5 and 6). Finally, a majority of students indicated that performing the genetic reporter assay increased their understanding of the assay and the use of genetic screens in general, but agreement on the two questions that addressed these goals (2 and 4) was not quite as high as on the other questions (just >70%).
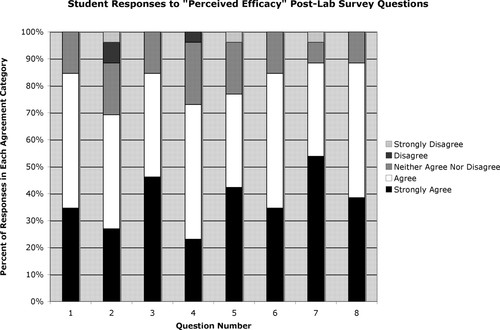
Figure 4. Five-point Likert scale responses to the eight questions of the postlab survey that addressed students' perceptions about the efficacy of the lab activities in meeting instructional objectives.
DISCUSSION
pp-α-F Western Blot Interpretation
In the pp-α-F Western blot, band b, which represents the full-length, unmodified pp-α-F polypeptide, is present at both temperatures in the sec61-1 cells (Figure 1A). This suggests that these cells have a partial defect in ER protein insertion even at the permissive temperature (25°C). When the students predict the results of the ss-HIS4 reporter assay, they are told to assume that 25°C is a fully permissive temperature for both mutant strains (i.e., no secretory defects), 30°C is semipermissive (i.e., partial defects), and 37°C is nonpermissive (i.e., full defects leading to nonviability). When they score their actual results from this assay, however, they discover that these assumptions are not completely true: the sec61-1 strain is not very tightly temperature sensitive, growing more slowly than WT even at 25°C, but still growing slightly at 37°C; sec18-1, in contrast, is extremely temperature sensitive, growing fine at 25°C, but growing poorly at 30°C and not at all at 37°C. Some students also observe a partially positive result in the ss-HIS4 reporter assay for the sec61-1 cells at 25°C (as well as the anticipated positive result at the semipermissive temperature of 30°C), corroborating the conclusion that sec61-1 has a partial secretory defect at all temperatures. It should be noted that, because sec61-1 grows more slowly than the other strains, the sec61-1 cell pellets were smaller, so the overall amount of protein loaded in the sec61-1 lanes of the blot shown in Figure 1A is lower than in the other lanes; if equivalent amounts of protein were loaded for all samples, it is likely that some of the glycosylated forms of the protein would be seen in at least the 25°C sample, because the viability of the strain at this temperature indicates that ER protein insertion cannot be totally blocked.
In the blot shown in Figure 1A, the two lower-molecular-weight forms of N-linked glycosylated pp-α-F show greater accumulation in the sec18-1 strain at 37°C than does the highest-molecular-weight form (compared with sec18-1 cells at 25°C). This could suggest that, when ER-to-Golgi transport is blocked, the buildup of secretory proteins in the ER overwhelms the ER glycosylation enzymes, such that proteins are not necessarily glycosylated on all possible sites. However, this particular result is not always seen; in other experiments it is the highest-molecular-weight form of pp-α-F (corresponding to the triply glycosylated protein) that predominates in the sec18-1 cells at the nonpermissive temperature.
The anti-pp-α-F antibody clearly exhibits substantial cross-reactivity with proteins other than pp-α-F, including one with a molecular weight of approximately 15 kDa, another at approximately 28 kDa, and many higher-molecular-weight proteins (Figure 1A). This seems to be a general feature of the pp-α-F sequence, because an antibody raised against the whole pp-α-F protein that we obtained as a gift from Randy Schekman (University of California, Berkeley) also showed significant cross-reactivity, although somewhat less than the antibody used in the blot in Figure 1A, which was raised against a 14 amino-acid peptide (data not shown). The cross-reactivity makes it essential to include a negative control sample of lysate from haploid MATa or diploid cells, neither of which produce α-factor (Figure 1A, leftmost lane).
Invertase Western Blot Interpretation
The intermediate-sized form of Suc2p-13myc that corresponds to the core glycosylated protein (band b in Figure 1B) is dramatically enhanced in sec18-1 cells at 37°C, reflecting the strong ER-to-Golgi trafficking defect in this strain at the nonpermissive temperature. Although the effect is less drastic at the permissive temperature (25°C), more protein is still seen in the intermediate-molecular-weight range and correspondingly less protein is seen in the highest molecular weight range for sec18-1 in comparison with the WT controls (Figure 1B). This suggests that sec18-1 cells have a slight defect in ER-to-Golgi trafficking in this strain even at the permissive temperature, something that would not be obvious from the yeast growth assay.
The sec61-1 cells show accumulation of the unglycosylated cell surface form of Suc2p-13myc at 25°C (Figure 1B, band c), but the cytosolic form of the protein that should show up just below this band (band d) is not visible in this sample. This is presumably due to the overall lower amount of protein loaded for the sec61-1 samples, as discussed for the pp-α-F blot. This may also explain why the myc-tagged invertase is nearly undetectable in the sec61-1 37°C lane and no higher-molecular-weight forms of invertase are detectable in the sec61-1 25°C lane. (Note that band c was visible in the sec61-1 37°C lane on longer exposures of the blot, but those longer exposure times resulted in overexposure and decreased resolution of bands in the other lanes.)
Because the anti-myc antibody is highly specific, the negative control lysate from cells that do not express and myc-tagged protein (Figure 1B, leftmost lane) is not essential for the interpretation of the data from this experiment; however, it is good training for the students to think about what controls would be appropriate for any experiment they do.
Experimental Considerations
Each of the two versions of the Western blot has its pros and cons. The high cross-reactivity of the anti-pp-α-F antibody complicates data interpretation in that version of the experiment and leads some students to question what they've been taught about the specificity of antibodies. However, the inclusion of a MATa-negative control sample ameliorates these problems. Lysates treated with endoglycosidase H to remove all oligosaccharides (perhaps prepared by the instructor) could serve as another set of controls. It is also possible that the specificity of the anti-pp-α-F antibody might be increased by using a preclearing strategy—preincubating the antibody with an acetone powder of MATa yeast cells, for example (Harlow and Lane, 1999)—but we have not yet tried this.
As mentioned above, the differences in the abundances of the three N-linked glycosylated ER forms of pp-α-F seen in the sec18-1 37°C sample compared with the sec18-1 25°C samples and the two WT samples also vary somewhat from one experiment to the next. This means that the sec18-1 ER-to-Golgi block may seem more or less striking depending on the particular experiment, but the data do always support the conclusion that the sec18-1 mutation causes an accumulation of normally secreted proteins in the ER. The mature, 3.4-kDa form of α-factor runs off the 12.5% polyacrylamide gels used in the pp-α-F Western blot experiment when the gels are run long enough to separate the three N-linked glycosylated ER forms of pp-α-F. If the mature form were visible, it would strengthen the interpretation of the sec18-1 data, because this form would be present in the WT cells but not in the sec18-1 37°C cells. Using 15% polyacrylamide gels might solve this problem. The major advantage of the pp-α-F Western blot is the clear visualization of the cytosolic form of the protein that is present in the sec61-1 cells.
The interpretation of the sec61-1 data are less clear in the invertase experiment due to the presence of the constitutively expressed cytosolic form of the protein, which is only 2 kDa smaller than the normally secreted form that becomes trapped in the cytosol in the sec61-1 strain. The gels must be run sufficiently long to separate these two cytosolic forms of invertase and, as seen in Figure 1B, the constitutive form is not always seen in the sec61-1 strain. Preparation of the cells used for the invertase experiment is slightly more involved than for the pp-α-F experiment, because the cells must be shifted from normal YPD medium to low-glucose YPD to induce expression of the secreted invertase, and a temperature shift of 3 h (rather than the 1 h used for the pp-α-F experiment) is required to see accumulation of the lower-molecular-weight bands in the sec18-1 strain. The major advantages of the invertase version of the Western blot are the commercial availability of the anti-myc antibody, the specificity of that antibody, and the clear distinction between the forms of invertase that accumulate in the sec18-1 strain and those that predominate in WT cells.
Ideally, equal amounts of protein from each cell lysate would be analyzed in the Western blotting experiment so that the amounts of each form of the chosen secretion substrate could be more accurately compared between samples. This would hopefully eliminate some of the ambiguities associated with both versions of the experiment. Equal protein loading could be accomplished in either of two ways: 1) measuring the total protein concentrations of the lysates and loading a set amount of protein; or 2) lysing the same number of cells for each sample. In practice, the first option is not very workable in the context of the current lab schedule; requiring the students to do a protein concentration assay before loading their gels would probably add an extra 45 min to the lab, causing it to exceed the allotted time period. Lysing equal numbers of cells would be more feasible. Currently, cell pellets for the entire class are prepared from large batch cultures by a member of our department's laboratory support staff, and culture densities are equalized by optical density measurements (600 nm) before the temperature shift; actually counting the cells using a hemacytometer at the end of the temperature shift and harvesting equal numbers of cells for each sample would likely provide improved results and would not be prohibitively labor-intensive. However, the inclusion of Ponceau S staining of the Western blot membranes before immunodetection allows students to assess the relative amounts of protein loaded in each lane and to use that information in their data interpretation.
Student Challenges
Both the ss-HIS4 reporter assay and the Western blot present significant intellectual challenges for the students. The experimental design of genetic assay is particularly confusing to most students when it is first presented to them, and at least one-third of students make some sort of error in predicting the yeast growth results in the homework that is assigned after lab 1. However, this confusion is usually resolved by week 2, after students score their results in consultation with their lab instructor. The scoring of the growth is itself a challenge, because the results are not always black and white: the exact density of a nega-tive control cell suspension spotted on the plates deter-mines whether that control truly shows no growth or if a slight “film” of growth is visible, so students must be reminded that they should use rational thinking in interpreting their data; for some students who prefer a clear “yes” or “no” answer, this subtlety—which is a common feature of the interpretation of genetic experiments—can cause uneasiness.
Requiring the students to make formal predictions of the growth of each strain in the reporter assay is essential to the pedagogical success of the experiment, because it helps the students to identify those strains that do not behave exactly as predicted when they score their actual results (i.e., the different temperature sensitivity profiles of the sec18-1 and sec61-1 strains). The students clearly appreciated this two-step process, as evidenced by their agreement with question 7 on the PE survey (“Making detailed predictions about the outcomes of the two experiments before seeing the results played a significant role in my learning process.”). Although the students are not required to predict the Western blot results as a homework assignment, they do this together as a whole class during the introduction to lab 2. Because many students have not interpreted Western blots before, and especially because of the cross-reactivity of the pp-α-F antibody, it is useful first to have the students predict the results in general terms, then to ask them to interpret an actual blot from a previous rendition of the experiment. Even having done this, most students still find interpreting their own results to be a challenging exercise, but they are able to do it with greater confidence than if they had not seen a sample blot beforehand.
Evaluating Learning Outcomes
The pre- and postlab KO surveys and the postlab PE survey have provided information that will be useful in improving student learning outcomes in future classes. They also provide a model for an assessment scheme that could be used to evaluate the efficacy of almost any pedagogical activity in either a lecture or a lab setting. Overall, the results from the surveys indicate that participation in the secretory lab module increased the students' knowledge about topics related to the lab and their confidence in that knowledge, and that they explicitly attributed those gains to the lab activities rather than to the lectures on the secretory pathway that they attended concurrent with the lab series.
The two questions on the PE survey that yielded the lowest percentage of students answering “Agree” or “Strongly Agree” were related to genetics (question 2: “Actually seeing the yeast growth results helped me better understand the ss-His4 reporter assay” and question 4: “This lab series improved my understanding of what is meant by a ‘genetic screen.’”). The relatively low agreement with question 2 (69%) was somewhat surprising, because one student had specifically remarked in class that she did not understand the assay until she saw the growth patterns on the plates. It is likely, however, that some students who did not agree with the statement in question 2 were those for whom the ambiguities of the data interpretation discussed above were particularly troubling. The results for question 4 were less surprising, because the students did not actually conduct a genetic screen during the lab module, but rather took advantage of a reporter gene that was previously used in a genetic screen (Deshaies and Schekman, 1987) to differentiate two known secretory mutants. Adding a class discussion of the paper by Deshaies and Schekman (1987) that first used the ss-HIS4 reporter might further the goal of increasing students' familiarity with yeast genetic screens as part of this lab module; such a discussion could occur while the students are running their gels or doing their Western transfers in week 2 or during the antibody incubations in week 3. Adding a paper discussion might also make more students feel that the lab module helped them to be “more comfortable interpreting blot figures in papers” (PE survey question 5).
The biggest disappointment on the KO survey was question 6, which asked whether the following statement was true or false: “A polyclonal antibody used in a Western blotting experiment will only recognize covalently modified forms of the protein if those forms of the protein were present in the antigen that was used to generate the antibody.” Only 18% of students chose the correct answer (false) in the postsurvey; this was a substantial increase from the 4% that answered correctly in the presurvey, but it was still rather low. The students were informed that the pp-α-F antibody was generated using a short peptide contained within the pp-α-F sequence (i.e., an unmodified form of the peptide), and they observed that it was able to recognize glycosylated forms of the protein, but they were apparently not able to connect these two pieces of information. In future classes, it will be important to help the students make this connection. The responses to question 4, which asked students to identify which statement about plasmids was untrue, were also disappointing; in the postsurvey, 57% of them chose the correct answer (“Plasmids can only be used to express genes in bacteria.”), but this was only slightly up from the 52% that answered it correctly in the presurvey. Because 25% chose the answer that was most obviously true (“Plasmids are circular pieces of DNA that can be engineered to contain genes of interest.”), rather than untrue, it is possible that some students misread the question.
The results for two other questions on the KO postsurvey warrant consideration for future courses: Question 3 (which concerned the use of conditional mutations to study the loss-of-function phenotype of an essential gene) and question 7 (which required detailed understanding of the variety of possible glycosylation events that may occur to a given protein). For both of these questions, the percentage of correct responses more than doubled from pretest to posttest (and for question 3 it actually increased sixfold), but the number of correct posttest answers was still <70%. Further emphasizing the fact that a knockout mutation in a essential gene produces a dead organism (not the best subject of study for dynamic processes) and that some proteins (like pp-α-F) are fully glycosylated in the ER, whereas others are further modified in the Golgi (like invertase) would help students better understand these concepts.
The KO and PE surveys were only administered to one class of 34 students in spring 2007 semester; however, anecdotal evidence gleaned from other, more general, surveys administered all 4 yr the module was taught (2004–2007) also supports the conclusion that the yeast secretory lab series is a successful teaching tool. These surveys included the college's standard Student Evaluation Questionnaire (SEQ), which students complete at the end of each course, as well as instructor-administered midsemester evaluations; both surveys included a question asking the students to identify the most valuable features of the course and aspects that could be improved. One laboratory instructor reported that, out of 30 students who responded to the SEQ in spring 2006, 25 of them identified learning new experimental techniques as the most valuable feature of the course, many explicitly mentioning the Western blot as being particularly useful. The same instructor noted that, in the 92 SEQs completed for her BISC 220 lab sections from 2004 to 2006, there was no negative feedback on the yeast lab series. Students SEQ responses included the following statements:
“The experiments we conducted were wonderful for reiterating the lecture material. While we may [have thought] we understood how a Western blot works, actually doing the procedure taught us more about [the] process.”
“This course provided a great overview of the functioning of the cell, and the laboratory did a great job of incorporating the ideas in lecture into a hands-on setting. The yeast screens were very interesting; I thought that [this lab] did a great job of showing us how the cellular transport system functions.”
These comments are representative of many others obtained from the SEQs and midsemester surveys, and they reflect consensus among the students that the yeast secretory pathway lab series made a positive impact on their overall learning in the course.
Summary
The Western blot experiments described here and the previously described ss-HIS4 genetic reporter assay (Vallen, 2002) function synergistically in a 3-wk laboratory series that gives students a hands-on understanding of two experimental approaches that have been instrumental in elucidating the molecular mechanisms by which proteins move through the endomembrane system. Based on the overall improvement seen in the number of correct responses to the KO survey postlab versus prelab, the lab module succeeded in increasing students' knowledge of the following: the various steps that make up the secretory pathway, the utility of yeast genetics for answering cell biological questions, and the way in which Western blotting can be used to track the “fate” of a protein within a cell. Furthermore, the responses to the PE survey indicate an overall high level of student satisfaction with the lab module in terms of its impact on their learning.
ACKNOWLEDGMENTS
I sincerely thank my BISC 220 coinstructors Tucker Crum and Gary Harris for their help in developing and implementing the lab and the evaluative surveys, the spring 2007 BISC 220 class for participating in the surveys, Padma Kannabiran for technical assistance with lab preparations, and Mary Allen and Kaye Peterman for helpful comments on the manuscript. Thank you also to Elizabeth Vallen and Vlad Denic for sharing yeast strains and plasmids and to Randy Schekman for providing the anti-pp-α-F antibody that was used in the initial development of the pp-α-F Western blot experiment. This article is dedicated to the memory of Glory Hom, a BISC 220 student who always brightened my day.