A Study of Rubisco through Western Blotting and Tissue Printing Techniques
Abstract
We describe a laboratory exercise developed for a cell biology course for second-year undergraduate biology majors. It was designed to introduce undergraduates to the basic molecular biology techniques of Western blotting and immunodetection coupled with the technique of tissue printing in detecting the presence, relative abundance, and distribution of ribulose-1,5-bisphosphate carboxylase in various plant materials. Pre- and postlab surveys indicated significant postlab gains in student understanding of all three lab techniques and relevant lecture topics. Additional postlab survey questions on student perception of the lab modules suggested that the laboratory exercises successfully met a series of pedagogical goals set by the instructors. The combination of these techniques provided a basis for quantitative and qualitative (visual) analysis of a biologically important enzyme and can be applied or modified readily to study other proteins and biological molecules in lab exercises for an introductory cell biology course or molecular biology course.
INTRODUCTION
Cell biology is an experimental science; therefore, the imparting of textbook knowledge and concepts resulting from rapidly accumulating scientific literature can be best complemented and enhanced through laboratory exercises. One of the challenges in teaching college-level introductory cell biology is offering undergraduate students laboratory experiences that are both hands-on and engaging, through well- thought-out and carefully designed experiments that make connections to lecture information. Because kinesthetic learning that emphasizes hands-on activities begins with young children and continues into the college laboratory setting (Tanner and Allen, 2004), it is arguably one of the most effective ways of student learning. The strategic selection and use of a variety of experimental techniques individually or in combination in labs benefit student learning by allowing them to gain understanding of scientific instrumentation and to master various techniques. Analysis of numerical and visual data as well as interpretation of experimental results develops critical-thinking skills. Overall, students get a chance to practice firsthand the process of doing science, and learn how scientists know what they know.
Biology majors at Truman State University are required to take cell biology in the fall of their second year. Students should have had 1 yr of introductory biology and a semester of inorganic chemistry, and most have had minimal experience performing independent research. We typically have three instructors teaching nine lab sections, with a total enrollment of 180–200 students each fall; this includes a significant portion of nonmajors (chemistry, exercise science, health science majors, and others). One of the long-standing goals of our cell biology laboratory is to introduce students to a variety of laboratory techniques that are used to study cells; this 3-wk laboratory exercise introduces the students to both Western blotting and tissue printing in the context of studying the relative abundance and distribution of the ribulose-1,5-bisphosphate carboxylase (Rubisco) enzyme in various plants and plant tissues.
In recent decades, plant materials have generally been neglected or underutilized in both precollege and college biology education by students and teachers alike (Wandersee, 1985; Hershey, 1993, 2002; Wandersee and Schussler, 2001). Because a considerable number of biology majors have an interest in pursuing careers in biomedical or health professions after completing their undergraduate education, they often fail to appreciate the important roles plants play in nature as well as in human and animal nutrition and medicine. Learning about and experimenting with plants would increase student understanding of plant function and importance, helping them to better appreciate the unique features of plant cell biology. Incorporating plant models in class experiments has unique advantages: they are maintained at low cost, readily available, easy to work with and dispose of, and have no concerns associated with animal experiments and dissections, such as the cost of care, regulatory constraints, and ethical issues (Hershey, 2005; Lally et al., 2007).
Rubisco is the most abundant soluble protein in the chloroplast and makes up 50% or more of all proteins in plant leaves; it is arguably the most abundant protein in the biosphere (http://4e.plantphys.net/article.php?ch=8&id=78, accessed 8 January 2009; Malkin and Niyogi, 2000). As a critically important enzyme to life on Earth, it catalyzes the chemical reaction by which inorganic carbon enters the biosphere and becomes incorporated into carbohydrates usable by most living organisms. Rubisco has a molecular mass of ∼560 kDa and consists of eight small (∼14 kDa each) and eight large (∼56 kDa each) subunits arranged as eight heterodimers (Malkin and Niyogi, 2000). In leaf chloroplasts, Rubisco “fixes” CO2 to a five-carbon acceptor (ribulose-1,5-bisphosphate) in the “Calvin cycle,” initiating its conversion to energy-rich molecules such as sucrose. The temporal (e.g., young vs. mature organ) and spatial (e.g., epidermis vs. cortex) expression of this important enzyme in various plants, tissues, and organs provides an interesting topic for investigative and inquiry-based student lab exercises.
Living cells constantly carry out a diverse array of metabolic activities. These processes form the basis of life, allowing cells to grow and reproduce, maintain their structures, and respond to various stimuli. An important aspect of cell biology research is to determine the spatial localization and temporal expression of specific molecules in tissues. An effective and simple technique for this purpose is tissue printing. Tissue printing is the art and science of visualizing cellular materials (e.g., macromolecules such as proteins and nucleic acids) in animal and plant tissues that are transferred to a receptive surface from a specific tissue (Varner and Ye, 1994). In this technique, a freshly and evenly cut section of any suitable biological specimen is pressed firmly on a nitrocellulose or nylon membrane, and the contents of the section are allowed to transfer to the membrane surface. The original tissue is then removed, leaving an “imprint” of the tissue anatomy on the membrane. Subsequent treatment of the tissue imprints by hybridization with an appropriate probe (e.g., RNA, DNA, or antibody) will indicate the location and abundance of specific cell components at the tissue level (Cassab and Varner, 1987; Taylor et al., 1993; Schopfer, 1994; Ruzin, 1999). Tissue print Westerns (for proteins) and Northerns (for RNAs) can be done with a variety of tissues and membranes (Varner and Ye, 1994). Although the resolution is somewhat less than that obtained from direct in situ hybridization on sectioned material, results can be obtained much more quickly and yield good anatomical details (Ruzin, 1999). Moreover, tissue printing can be readily mastered by inexperienced laboratory personnel, in our case, second-year undergraduates.
Many useful techniques, such as electrophoresis and Western blotting, among others, are widely used in cellular and molecular biology research. The combined use of these methods helps researchers to gain deeper insight into a molecule's localization, abundance, structure, and function. In this lab exercise, we attempted to achieve the following objectives: 1) convey the necessary technical knowledge to perform electrophoresis and Western blotting; 2) teach students the basics of image analysis and a mathematical method to analyze the result in Excel (Microsoft, Redmond, WA); 3) introduce tissue-printing techniques in a fun, inquiry-based manner; and 4) expose students to the much neglected discipline of plant cell biology. We used celery petiole because of its large size and ease of handling to localize Rubisco on its cross-sections; however, other plant tissues can and have been used, including broccoli, carrots, green beans, and green and red tomatoes. The nutritional value and potential medicinal use of celery provides an interesting subject for students to explore on their own through online literature research during the “downtime” (i.e., incubation steps) in the lab.
MATERIALS AND METHODS
Plant Material
Several plant species were used in this lab, including celery and snap pea purchased from a local grocery store, and green bean and onion obtained from our own teaching and research greenhouse. In prelab preparations, leaf and petiole samples from young and mature celery were taken, along with leaves from bean and onion plants, and snap pea pods and onion bulbs. For isolating and detecting Rubisco from various plants using Western blotting, fresh tissues were homogenized with pestles in prechilled mortars on ice in 1 ml of cold homogenization buffer (100 mM Tris, pH 7.5, 10% sucrose, 5 mM sodium EDTA, and 5 mM sodium EGTA) for each gram of tissue. Preparation of tissue homogenates was conducted by the instructors, who then performed a Lowry protein assay (Lowry et al., 1951) to determine protein concentrations. The appropriate amount of aliquots (20 μl containing 20 μg of protein) to be used by students in Western blotting was dispensed before lab. In a separate introductory lab exercise before starting the lab series of this study, students also performed a Lowry protein assay on the same tissue homogenates and compared their results with those obtained by the instructors.
For localizing tissue distribution of Rubisco, celery stalk was used. All steps were carried out at room temperature.
Equipment, Chemical Reagents, and Procedures
Each of the following three procedures makes up a 1-wk lab module and can be completed in a 2-h 50-min lab session. With fresh supply of reagents, the one-time average cost per student was approximately $5 for each lab. However, because the quantity of many of the reagents is enough to support several semesters' use, the actual cost of the lab series becomes lower over time. Because of the large size of our laboratory sections (20–24 students), students worked in groups of four to execute each of the lab modules.
Electrophoresis and Western Blot (Week 1)
Students loaded plant tissue samples (20 μg of total protein in 20 μl), a Rubisco standard prepared from spinach (0.5 μg in 20 μl; Sigma-Aldrich, St. Louis, MO), and EZ-Run protein molecular mass standard (Thermo Fisher Scientific, Waltham, MA) onto a precast polyacrylamide gel (PAGEgel 4–20% TEO-CI SDS; Fisher Scientific, Hanover Park, IL) using Hamilton syringes (Hamilton, Reno, NV). The gels were run at approximately 80 milliamps for 30–40 min in electrophoresis buffer (PAGEgel SDS running buffer, Fisher Scientific) by using a Novex Mini-Cell II electrophoretic unit (Invitrogen, Carlsbad, CA). Proteins were then transferred to Protran nitrocellulose transfer membrane (Whatman, Dassel, Germany) in McFarland's electroeluting buffer (12 mM Tris base, 96 mM glycine, and 20% methanol) at <25 V for ∼45 min. Students were cautioned about a potential pitfall before beginning this procedure, which may occur at the time of assembling the “gel-nitrocellulose membrane sandwich”: if it was oriented incorrectly in the blot module with respect to the direction of the electric field, the proteins would not be transferred to the nitrocellulose membrane, and the subsequent immunodetection step would not yield any meaningful signal. The blotted and air-dried nitrocellulose membrane was placed in a ziplock bag and stored at −20°C until immunodetection of Rubisco.
Immunodetection of Rubisco (Week 2)
To begin Rubisco immunodetection, the nitrocellulose blot was placed in 10 ml of blocking solution (5% nonfat dry milk in 24.8 mM Tris, pH 7.4, 150 mM NaCl, 2.7 mM KCl, and 0.5% Tween 20 [TBST]) and rocked gently on a rotary shaker or platform rocker for 30 min. This step was begun before students arrived to save time. The students then washed their blots four times for 5 min each with TBST and incubated them for 1 h on a shaker-rocker with polyclonal rabbit anti-Rubisco antibody (anti-Rubisco large subunit global antibody, diluted 1:6000; Agrisera, Vännäs, Sweden). After four 2.5-min washes with TBST, the blot was incubated with goat anti-rabbit immunoglobulin G (IgG) polyclonal antibody conjugated to alkaline phosphatase (diluted 1:10,000; Sigma-Aldrich) for 30 min on the shaker. After four 2.5-min washes, 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium chloride (BCIP/NBT) chromogenic substrate solution for alkaline phosphatase (Sigma-Aldrich) was added, and the blot was allowed to develop, which generally took no longer than 15 min. The blot was then rinsed three times in water for 2 min each. The developed blot was allowed to dry on a clean piece of filter paper and stored in a ziplock bag at −20°C. The Western blot image of protein bands was captured with a scanner (HP Scanjet 4850; Hewlett Packard, Palo Alto, CA) or other image capture system, and protein band positions of each of the 10 proteins in the molecular mass standard as well as Rubisco from plant tissues were determined using the length measurement tool in ImageJ software (ImageJ 1.34s; http://rsbweb.nih.gov/ij/) or manually with a ruler. The molecular mass of Rubisco from experimental samples was estimated by graphical analysis through comparing migration distances to those of protein markers of known sizes. Teaching of other image analysis skills for quantifying the intensities of bands is planned for future labs.
Tissue Printing and Western Blot Analysis (Week 3)
This procedure is adapted from the tissue printing procedure by Ruzin (1999). Students prepared a fresh section of celery petiole by cutting with a clean single-edged razor blade to approximately 0.5 cm in thickness. Students then pressed the newly cut surface of the celery petiole firmly and evenly onto Protran nitrocellulose membrane, which had been placed on top of six layers of Whatman no. 1 paper, for 15–20 s. Care was taken not to squash the tissue. Multiple prints from different freshly cut sections were made on a single piece of nitrocellulose membrane, and students were instructed to make a drawn record of the tissue print pattern.
To detect the presence and location of the Rubisco enzyme, the nitrocellulose membrane was washed in washing buffer 1 (0.1 M Tris-HCl, pH 8.0, 0.05% sodium azide, and 0.3% Tween 20) for 5 min, followed by a 10-min incubation in blocking buffer (0.1 M Tris-HCl, pH 8.0, 0.05% sodium azide, 0.3% Tween 20, 0.25% bovine serum albumin, and 0.25% gelatin). The membrane was then incubated for 1 h on a rotary shaker with polyclonal rabbit anti-Rubisco antibody (diluted 1:1800 in blocking buffer; Agrisera). After this step, the membrane was washed four times with washing buffer 1 for 1 min each and then incubated in goat anti-rabbit IgG polyclonal antibody conjugated to alkaline phosphatase (Sigma-Aldrich), diluted 1:1000 in blocking buffer, for 30 min on a rotary shaker. The membrane was then washed once in washing buffer 1 for 1 min, twice for 1 min each in washing buffer 2 (0.1 M Tris-HCl, pH 8.0, 0.05% sodium azide, 0.3% Tween 20, and 0.05% SDS), and one final time in washing buffer 1 for 1 min. Next, the membrane was equilibrated with AP buffer (0.1 M Tris-HCl, pH 9.5, 0.1 M NaCl, and 5 mM MgCl2) for 1 min, followed by addition of BCIP/NBT chromogenic substrate solution for alkaline phosphatase (Sigma-Aldrich). Purple color occurred where the protein of interest, i.e., Rubisco, was localized, and the color development was complete within 10 min. To stop the reaction and avoid a dark background, the membrane was placed in washing buffer 3 (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) for 5 min, followed by washing in distilled H2O for 5 min. Finally, the membrane was thoroughly air-dried.
Observation of the final tissue print was carried out visually, under a dissecting microscope, or a combination. The nitrocellulose membranes with tissue prints were scanned and saved as electronic files, and then they were stored between two pieces of Whatman no. 1 paper in a ziplock bag at −20°C. Students could then print out the electronic image file and include it in their lab notebook or lab report.
Assessment of Learning Objectives
Prelab and postlab surveys were given to the students to assess the success of learning objectives. The prelab survey served as a baseline test aimed at measuring student knowledge before learning any of the lab techniques, concepts, or specific factual information and was given 3 wk before the three lab modules began. It consisted of 18 multiple-choice questions, including both those that were relevant to the labs and several nonspecific control questions, so that the target lab concepts were not so obvious to the students. The questions were grouped into four general categories: 1) knowledge learned from previous and current course work; 2) specific knowledge on Rubisco, the photosynthetically important enzyme that was to be studied in the three labs; 3) specific knowledge on techniques, i.e., the understanding of how each of the three laboratory techniques works; and 4) the ability to consider the applicability of the taught techniques in research. Category 1 served as a control and included four questions, one of which the students should have had some prior knowledge about, two concerned topics taught in other lab exercises, and the last on a subject taught in the lecture but not studied in any labs. The postlab survey was given in the week after the completion of the three lab exercises, and responses were used to assess specific goals. It included the same 18 questions from the prelab survey, as well as seven additional questions that evaluated student perception of their lab learning experiences. All surveys were completed during the lab to ensure uniformity in testing conditions. Data presented are based on responses from 183 (prelab) and 185 (postlab) students in all nine lab sections (exempt from review by Truman State University's Institutional Review Board). Depending on the instructor, each student was required to either write up an individual lab report that followed the format of typical journal articles or to include a write up in his or her lab notebook (a separate lab write up for each week of the lab); both formats included an introduction to techniques, results, and data interpretation. This was done in addition to the lab surveys.
Multivariate Hotelling T2 test (Hotelling, 1931) was used to compare pre- and postlab survey scores for each of the categories described above. Hotelling's T2 is a multivariate extension of the Student's t test. In a t test, differences in the mean response between two normally distributed populations are studied. T2 is used when there are two or more response variables with multivariate normality assumption, although it can be used when there is only one response variable. In our analysis, estimated proportions approximately followed normal distribution with a large sample size of >180 individuals. Based on results from the T2 test, a one-sided independent t test was also used to compare the pre- and postlab scores for each question in any given category. All statistical tests were conducted with the statistical package SPSS version 16 (SPSS, Chicago, IL).
RESULTS
Each group of students produced their own Western blot and tissue print for Rubisco. The results shown here belong to one group and are representative of the class data.
Detection of Rubisco through Electrophoresis and Western Blot
Figure 1 shows the presence and relative abundance of Rubisco from various plants; and in celery, young and mature leaves and petioles can be compared. Students were asked to interpret what they found on their blot, including qualitative differences in band density. By examining their blots, students were able to report that Rubisco was present at varying levels in all of the plant samples with green tissues and absent from the white onion bulb. Green bean leaves contained the most Rubisco, followed by snap pea pod, green onion leaves, and celery. Young celery leaves had relatively less Rubisco than mature leaves, but young celery petioles had slightly more Rubisco than mature petioles.
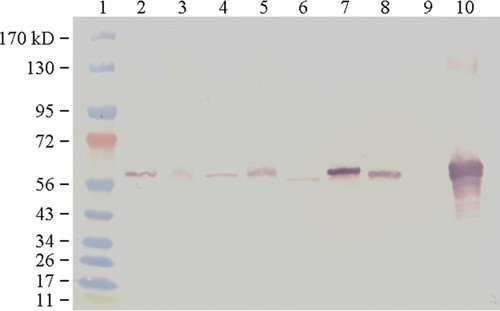
Figure 1. Representative Western blot of Rubisco produced by students. The two left-hand most lanes represent molecular marker proteins (lane 1) and Rubisco standard (lane 2). The remaining lanes show Rubisco extracted from various plants (from left to right): young celery leaf (lane 3), young celery petiole (lane 4), mature celery leaf (lane 5), mature celery petiole (lane 6), snap pea pod (lane 7), onion leaf (lane 8), onion bulb (lane 9), and bean leaf (lane 10).
Estimation of the Molecular Mass of Rubisco Large Subunit
Students were then asked to estimate the molecular mass of Rubisco large subunit from the Western blot. They first measured the migration distances of each of the 10 marker proteins in the molecular mass standards and then correlated the distances with the known molecular masses. After measuring migration distance of the Rubisco in the various plant samples, they calculated the molecular mass of Rubisco from the mathematical function describing the correlation (Figure 2). The calculated molecular mass of the large subunit of Rubisco from various plants used in the exercise is shown in Table 1. The variation in molecular mass compared with the Rubisco standard prepared from spinach was within 5% for all of the plant materials used.
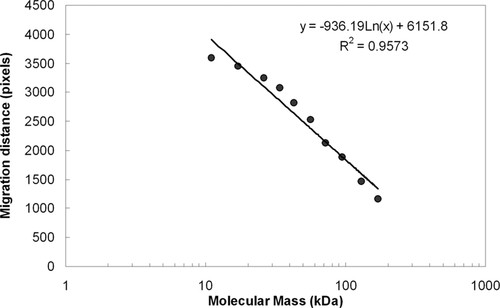
Figure 2. Migration distances of marker proteins measured from the Western blot shown in Figure 1 and plotted against the logarithm of the molecular mass.
| Sample | Migration distance (pixels) | Calculated molecular mass (kDa)a | % difference from Rubisco standard |
|---|---|---|---|
| Rubisco standard (from spinach) | 2442 | 52.6 | |
| Young celery leaf | 2442 | 52.6 | 0 |
| Young celery petiole | 2436 | 52.9 | 0.6 |
| Mature celery leaf | 2430 | 53.3 | 1.3 |
| Mature celery petiole | 2490 | 50.0 | −4.9 |
| Snap pea pod | 2412 | 54.3 | 3.2 |
| Onion leaf | 2448 | 52.3 | −0.6 |
| Bean leaf | 2406 | 54.7 | 4.0 |
Localization of Rubisco through Tissue Printing
Figure 3 shows the distribution of Rubisco in tissues of young and mature celery petioles. At the beginning of the lab, students were asked to predict where Rubisco would be found on a cross-section of the celery petiole, and whether they would expect differences in Rubisco distribution and content between young and mature petiole. Similarly, students hypothesized whether celery that had been stored under dim light for several days would have an altered level of Rubisco. They would then describe the localization of Rubisco from the tissue prints they prepared. It can be seen that Rubisco was present in both young and mature petioles, and was mostly accumulated in the epidermal layer (i.e., the green tissue) as well as the vascular bundles (i.e., tissues for transport of water and minerals from the root to the shoot, and photosynthetic products (carbohydrates) from shoot to other organs). Postharvest celery kept under dim light led to a reduction of Rubisco content in the petiole (Figure 3).
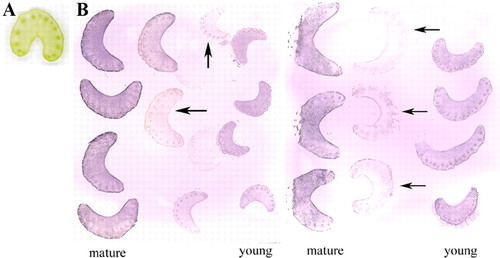
Figure 3. (A) Scanned image of a cross-section of fresh celery petiole. (B) Examples of student-prepared tissue prints showing distribution of Rubisco on cross- sections of celery petioles. The print was produced by pressing the cut surface of celery petioles on a piece of nitrocellulose membrane, which was then incubated with anti-Rubisco primary antibody, followed by a secondary antibody conjugated to alkaline phosphatase. Purple color indicates the presence and location of Rubisco. Cross-sections shown are from young and mature celery petioles. Arrows indicate prints from celery petioles kept in water and under dim light for several days before prints were made, showing decreased Rubisco content.
Assessment of Student Learning
The objectives of this study were multifaceted. We attempted to teach students the principle and techniques of electrophoresis and Western blotting, the basics of image analysis, and a mathematical method to quantify protein molecular mass in an Excel spreadsheet. We also sought to introduce tissue printing technique to the students in a fun and inquiry-based manner, and expose them to the use of plant materials in cell biology study. Pre- and postlab knowledge and opinion surveys were used to assess the extent of student learning and to evaluate the pedagogical value of the multiweek lab exercise. In total, 183 and 185 students participated in the pre- and postlab surveys, respectively. If a student missed the prelab survey, the student was excluded from the postlab quiz, although there were a few included inadvertently. Student performance improved significantly in all new knowledge categories, as measured by the multivariate hotelling T2 test (Table 2); furthermore, a one-tailed independent t test shows that the percentage of students answering each question correctly within a knowledge category increased significantly in the postlab survey, with a few exceptions (Table 2; see below), indicating improved learning of specific knowledge related to the lab exercises, whereas no significant change in the survey results was found in the question designed to assess their prior knowledge (Table 2).
| Question category | Question no. | % correct prelab (n = 183) | % correct postlab (n = 185) | Change (post–pre) | P value, multivariate hotelling T2 test | P value independent t test |
|---|---|---|---|---|---|---|
| Rubisco: the enzyme studied in the three labs | 3 | 37 | 79 | 42 | 0.000 | 0.000 |
| 5 | 28 | 57 | 29 | 0.000 | ||
| 6 | 30 | 59 | 29 | 0.000 | ||
| 8 | 44 | 85 | 41 | 0.000 | ||
| Technique 1: SDS-PAGE | 9 | 32 | 87 | 55 | 0.000 | 0.000 |
| 10a | 68 | 68 | 0 | 0.363 | ||
| 11 | 28 | 50 | 22 | 0.000 | ||
| 12 | 31 | 85 | 54 | 0.000 | ||
| Technique 2: Western blot | 13 | 30 | 88 | 58 | 0.000 | 0.000 |
| 14 | 35 | 48 | 13 | 0.002 | ||
| Application of techniques 1 and 2 | 16 | 26 | 33 | 7 | 0.032 | |
| Technique 3: tissue printing | 15 | 36 | 45 | 9 | 0.011 | |
| Application of technique 3 | 17a | 74 | 57 | −17 | 0.004 | |
| 18b | 16 | 81 | 65 | 0.000 | ||
| Concurrent lecture topics | 2c | 87 | 90 | 3 | 0.000 | 0.002 |
| 4c | 54 | 77 | 23 | 0.000 | ||
| 7c | 18 | 22 | 4 | 0.128 | ||
| Previous knowledge (control) | 1 | 94 | 90 | −4 | 0.183 |
In the postlab survey, 80% of all students responded affirmatively to the helpfulness of the three lab modules in their understanding of the lab techniques (Figure 4); this self-assessment was supported by their improved performance on the postlab survey (Table 2). Nearly 60% of students reported learning the tissue printing technique in a fun way; 52% enjoyed working with plants and 50% expressed excitement at seeing the tissue printing result; 65% of students said they could envision applicability of the techniques, and 51% became more comfortable at using Excel to process experimental data (Figure 4). With respect to knowledge retention, 28% of students said they were able to recall essential information on all three labs independently; 54% said they needed some help, and only 6% said they needed much help.
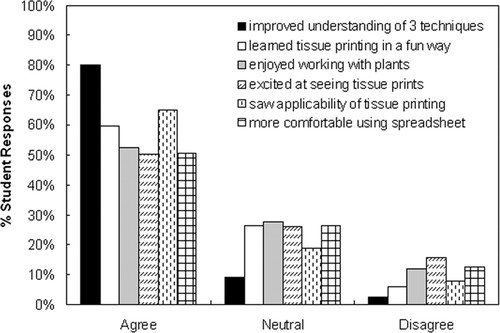
Figure 4. Student perception of their learning experiences on the three lab modules. “Agree” includes both “Strongly agree” and “Agree,” and “Disagree” includes both “Strongly disagree” and “Disagree.”
The survey questions we developed were based on the multiple-choice answer format, and two of the 17 questions had a correct answer that included all of the given choices, i.e., in the form of “all of the above” (Table 2, questions 10 and 17). By the time ∼30% of the 183 students completed the prelab survey, we found that approximately 70% of students had chosen the correct answer. This was a surprisingly high percentage considering that the two questions concerned completely new material to most students. Consequently, we added a follow-up question (question 18) to one of the two questions (question 17), asking for students to rank their level of confidence in their chosen answer for question 17 (i.e., knew the answer, reasoned, guessed, other) and gave the modified survey to the remaining students, none of whom selected “knew the answer” even when they got the correct answer. Interestingly, on the postlab survey, there was no change in the percentage of correct response to the first of the two questions (question 10) but a 17% decrease in the percentage of correct response to the second question (question 17), which had a follow-up question asking for confidence of choice. However, there was a significant 65% increase in the number of students who said they knew or reasoned the answer to question 17 on the postlab survey (Table 2).
DISCUSSION
In general, the three weekly lab modules fit nicely together and formed a logical sequence in helping students to learn and understand the techniques of electrophoresis, Western blotting, immunodetection, and tissue printing. During the process, they also learned how to extract qualitative (i.e., relative abundance of Rubisco and its tissue distribution) and quantitative (i.e., molecular mass estimation of Rubisco large subunit) information from the results through image analysis and mathematical manipulation and to build connections between these results and what they represent in plant tissues. Postlab responses indicated that 80% of the students reported improved understanding of the lab techniques (Figure 4). Because each lab module focused on one technique, students had sufficient time to become familiar with each set of protocols during the lab period. The lapse of one week in between each lab session required students to recall essential background information and objectives of what they attempted to achieve in the previous lab to see the logic of continuity throughout. This repetition helped strengthen the retention of important concepts they learned each step of the way, as evidenced by improved performance on their postlab compared with prelab surveys (Table 2).
Students often experienced difficulty with math in the calculation of molecular mass of the Rubisco. They were instructed to enter the measured migration distances of the molecular mass markers and the molecular mass values into the Excel spreadsheet, plot the distance against the logarithm of the molecular mass of the marker proteins, find the trend-line in the form of a linear equation [i.e., Y (migration distance) = k × log (molecular mass) + b], get the R2 value, and see how well the equation describes the log-linear relationship. They then calculated the molecular mass of the large subunits of Rubisco for various plant samples by entering into the equation the migration distance of the Rubisco, and solving for the molecular mass. The struggle often occurred at the last step of solving for molecular mass from the equation of log (molecular mass) = known value, the answer of which is obtained by converting to the exponential function (i.e., molecular mass = e raised to the power of the known value). In addition, students showed varied proficiency in data processing with Excel. Many students were preoccupied with using a handheld calculator to process data, until they saw the ease and speed of repetitive calculation in the spreadsheet and learned how to use mathematical functions and create charts in Excel to visually present experimental data. This 3-wk lab module was the last one among a few other labs in which students had an opportunity to learn the use of Excel. Fifty-one percent of students reported that they became more comfortable using a spreadsheet, whereas 26% had a neutral response, and 12% expressed difficulty with Excel (Figure 4). We believe that helping students in applying math to analyze biological data and in the transition from using a calculator to the more sophisticated data-processing software is an added benefit to students' learning experience, an aspect in undergraduate biology education that was recommended by the National Research Council (2003).
The tissue printing module offered an enjoyable lab experience for nearly 60% of students, and 52% of students enjoyed working with plants (Figure 4). Celery, the plant material used for the prints, is readily available and hardy to handle. Each student in a group was encouraged to make a print, with multiple prints made on each piece of nitrocellulose membrane; this increased the chance of getting a successful print. The physical mapping and visualization of protein distribution in tissues from tissue prints proved to be exciting for half of all the students.
One potential problem with these procedures was the downtime during the longer steps in the procedure. A strategy we applied to use these incubation times was for students to analyze, interpret, and discuss their data collected in the previous lab module. Students were also encouraged to conduct literature searches on the nutritional value and medicinal use of celery, and required to observe and record the anatomy of celery petioles under a dissecting scope. In one of the instructors' three lab sections, students discussed and debated their prediction on the tissue location of Rubisco, and whether young and mature petioles would differ in Rubisco content. They generally agreed that they would see Rubisco in the green epidermal tissue but were surprised to see its presence in the inner vascular tissue as well. They disagreed over possible differences in Rubisco content between young and older petioles, citing growth rate difference and location of young versus older stalks relative to each other within a bundle and whether the light source could be a contributing factor. Although the tissue prints they generated did not answer all of these questions equally well, they clearly benefited from this inquiry-based approach.
Student learning of the lecture material and their understanding of lab techniques benefited significantly from the hands-on activities in the lab. In addition to the significantly enhanced postlab survey performance on questions specifically related to the three lab modules reported here (Table 2), students scored well on two other lecture topics that were also associated with lab exercises. However, they did not show improvement in knowledge on another photosynthetically important enzyme (i.e., phosphoenolpyruvate carboxylase) that was either addressed in the lecture or included in the text reading assignment, but to which no lab activity was devoted (Table 2, concurrent lecture topics).
The development of a good assessment questionnaire requires much care and skill to avoid potential problems. The survey questions we developed were of multiple-choice format and can be considered to be a generally effective and reliable measurement of student learning overall; however, we found it important to exclude inclusive correct answers in the form of “all of the above,” which increased the chance of student guessing the correct answer and risked greater bias in student assessment (Table 2, questions 10 and 17). Still, remedy is possible for more in-depth analysis and insight into student response to questions with such answers by including a follow-up question that probes students' confidence in their response.
In examining students' ability to apply the techniques they learned, we found that although there was improvement in student knowledge on how the techniques might be applied beyond the class studies, it was a relatively small gain compared with understanding the technical details and facts of the particular molecule under study (Table 2, application vs. other categories). Considering that most of our students did not have more advanced knowledge in biochemistry, organic chemistry, or both, we believe that their ability to apply or transfer a technical approach beyond the immediate experience will be improved as they gain more knowledge on the physical and chemical properties of biological molecules from other course work.
In summary, the results of the assessment surveys show that we achieved the stated learning objectives of each of the lab modules, and we believe that they can be applied or modified as a feasible and beneficial introductory cell biology lab exercise for undergraduates in learning important cell biology techniques and their potential applications.
ACKNOWLEDGMENTS
We thank Deborah Hudman for prepping and setting up all labs; Dr. Julie Lochbaum (Truman State University's Center for Teaching and Learning) for helpful discussions and advice on the development of assessment questionnaire; and Truman State University students in Biol 200, fall 2008, for hard work and willingness to share responses for this study.



