Relational Databases: A Transparent Framework for Encouraging Biology Students To Think Informatically
Abstract
We discuss how relational databases constitute an ideal framework for representing and analyzing large-scale genomic data sets in biology. As a case study, we describe a Drosophila splice-site database that we recently developed at Wesleyan University for use in research and teaching. The database stores data about splice sites computed by a custom algorithm using Drosophila cDNA transcripts and genomic DNA and supports a set of procedures for analyzing splice-site sequence space. A generic Web interface permits the execution of the procedures with a variety of parameter settings and also supports custom structured query language queries. Moreover, new analytical procedures can be added by updating special metatables in the database without altering the Web interface. The database provides a powerful setting for students to develop informatic thinking skills.
INTRODUCTION
The ongoing genome projects have led to the creation of data sets of a size and complexity that is new to the field of biology. Students of biology now face the challenge of building analytical skills to begin to understand these new kinds of data. In addition to being able to think about biological processes from molecular to organismal and community levels of organization, tomorrow's biologists will also benefit from new bioinformatic ways of thinking derived from the fields of computer and information sciences (Kanehisa, 2000; Murray, 2000).
The current development of the new genomic-scale data sets leads to two natural questions: What are the best ways to represent large-scale data sets? What are the best ways to analyze large-scale data sets? The challenge in the field of bioinformatics is to learn to think in terms of large data sets (e.g., not one gene at a time, but thousands of genes) and to develop analytical approaches to extract biological information from large data sets. Genomic-scale data sets are being developed for many kinds of biological information, including gene expression, protein structure and function, and DNA and protein sequence data. We can think of genomic sequence data sets as defining special sets of sequences (sequence spaces) that can be contrasted with sets of random sequences (random sequence space). By identifying the constraints that define the sequence spaces, we hope to discover potential properties or patterns in the biological processes underlying the data sets that can then be tested experimentally. The central goal is to extract biological meaning through informatic analysis of the data, but how can we represent these sequence spaces, and how can we identify the constraints? We discuss below how the intrinsic structure and standard analytical power of relational databases provide a powerful setting for informatic analyses of genomic and related data sets.
Ideally, we would like to have a transparent framework that provides biology students the excitement of exploring large data sets, looking for underlying patterns, and analyzing the data without the need for sophisticated programming expertise. This framework should facilitate the development of new analytical approaches and ways of understanding complex genomic information. As we describe here, relational databases provide just such a setting. Indeed, their intrinsic structure provides a powerful setting for informatic analyses of genomic and related data sets that can be readily used by students.
In this article, we discuss our use of a Drosophila splice-site database to encourage students to think informatically. In the case study, we present examples of informatic analysis with the database to illustrate its analytical power. A full description of this analysis is published elsewhere (Weir and Rice, 2004). In the sections on Student Experiences and Assessment, we discuss student experiences when being introduced to informatic ways of thinking through use of the database. Although only used once, as a component in a small-enrollment class, this initial feedback indicated that hands-on experience manipulating large data sets provides students from biology and computer science backgrounds with a valuable experience in starting to think informatically. In Appendix A, we discuss general properties of relational databases that make them an ideal framework for representing and analyzing large biological data sets. We also discuss the use of stored procedures that permit more complex analyses of data sets. In Appendix B, we describe the design of the Drosophila splice-site database, focusing on how its design facilitated adding new analytical methods as our analysis of splice sites proceeded. We note that the essence of the database design is captured in Figure 1, and the details presented in these appendices are not essential for our discussion. However, the reader will find it useful to review the discussion of databases in the appendices—how data is stored, retrieved, and analyzed—in assessing the value of databases in bioinformatics curricula.
Figure 1. Data table schema. Schema for data tables in our Drosophila splice-site database (see Appendix B for detailed discussion).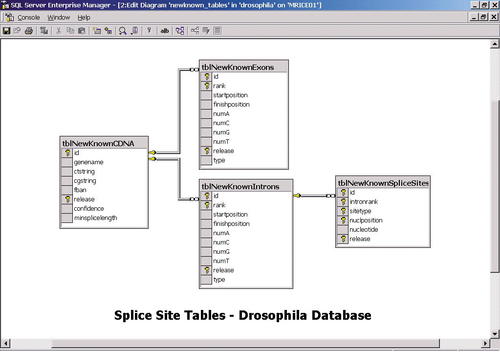
We emphasize that our primary goal in introducing students to relational databases of biological data sets was to nurture informatic thinking skills. Learning programming languages, principles of database design, and writing stored procedures were not primary goals—these skills can be developed in other courses. By using versatile stored procedures available in our splice-site database, biology students could explore and informatically analyze a large biological data set without being stifled by the mechanics of programming. Figure 2 shows examples of the kinds of genomic-scale questions that students were able to address (see section on Student Term Projects). In contrast to working with many biological databases that are typically queried one element at a time, students could address questions about genomic-scale relationships within populations of sequences.
Our experience indicates that genomic databases provide a useful context for introducing students to informatic ways of thinking through different analytical approaches. These approaches include: information-theoretic analysis of consensus sequences (see Development of a Genomic Database), progressive partitioning of sequence spaces (see Development of a Genomic Database), construction and assessment of algorithms for biological analysis and processes (see Assessing Data Quality and Algorithm Design), testing of genomic data set relationships (see Bioinformatic Analysis without Programming, and Designing Queries), and exploration of alternative data representations to answer different questions (see Student Term Projects). These approaches allow students to become comfortable with manipulating large biological data sets in a variety of structured ways that can lead to new insights into the corresponding biological processes.
Figure 2. Genomic scale questions for student term projects. Students used our Drosophila splice-site database to address these questions in their term projects in our bioinformatics course.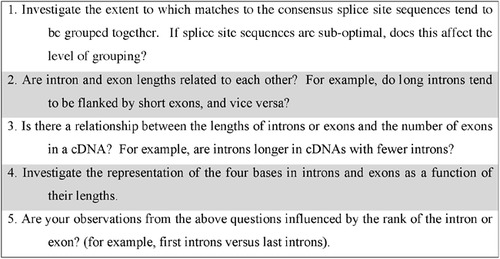
DEVELOPMENT OF A GENOMIC DATABASE: A CASE STUDY
We have adopted the ideas discussed in the introduction to develop a relational database for Drosophila splice sites (see Appendices A and B). We embarked on this project for two reasons. The primary reason was that we wished to analyze splice sites on a genomic scale to gain insights into spliceosome function (Fields, 1990; Lim and Burge, 2001; Mount et al., 1992; Stephens and Schneider, 1992). In addition, however, we wished to develop a genomic-scale data set for students in our undergraduate bioinformatics course to allow them to gain experience using analytical approaches with a large data set. In this section, we introduce the splice-site database with examples of the useful analyses it made possible (Weir and Rice, 2004) that formed the basis for components of our bioinformatics course, discussed in the sections on Student Experiences and Assessment.
The database was developed using the Microsoft SQL Server 2000 database management system and stores data about exons, introns, and splice-site regions. These types of data were computed using a custom algorithm that matches transcript (cDNA) sequences (Stapleton et al., 2002) with genomic DNA sequences (downloaded from the Berkeley Drosophila Genome Project). The splice-site data were stored in database tables as described in Appendices A and B. The data organization is summarized in the database schema (Figure 1, see Appendix B). A collection of stored procedures was developed to perform a variety of specific data analyses, including the computation of information content at nucleotide positions near splice sites, frequency distributions of nucleotides, and intron-length distributions (see following discussion). In addition, the database contains special metadata tables that support a generic Web interface written in the Perl language (Appendix B). By updating these tables, new stored procedures can be made available to users without making any changes in the Web interface.
The relational database greatly facilitated the analysis of sequence conservation at splice sites—an example of a constrained sequence space (see Introduction). In particular, several of the stored procedures permitted efficient testing of different specialized contexts that enhance sequence conservation. Once features that appeared to influence splice function were identified (such as intron length), they were added as parameters in procedures to test other contexts. The data set needs to be sufficiently large (in our case, 10,057 introns) so that when it is partitioned on the basis of a parameter such as intron length, the resulting subsets are of reasonable size. Within these subsets of the data, we can look for other parameters that also influence splice function. Clearly, very large data sets are crucial for this approach, which will become increasingly useful as more large genome data sets become available.
The following example illustrates how one of the stored procedures can be used to calculate information at nucleotide positions near donor splice sites. Information, as defined by Schneider (Stephens and Schneider 1992), provides a sensitive measure for quantifying sequence conservation at a given position in an alignment. It is defined by the quantity  where fA,..., fT are the frequencies of each nucleotide at the given position, and c is a correction factor that depends on the number of splice sites being considered (see Weir and Rice, 2004). Information is tied to the notion of uncertainty, and the amount of uncertainty refers to how many decisions are required to specify the nucleotide at a particular position. If all four nucleotides can be present at a position, then two binary decisions (2 bits) are sufficient: Is the nucleotide a pyrimidine or purine? If the latter, is it an A or G?
where fA,..., fT are the frequencies of each nucleotide at the given position, and c is a correction factor that depends on the number of splice sites being considered (see Weir and Rice, 2004). Information is tied to the notion of uncertainty, and the amount of uncertainty refers to how many decisions are required to specify the nucleotide at a particular position. If all four nucleotides can be present at a position, then two binary decisions (2 bits) are sufficient: Is the nucleotide a pyrimidine or purine? If the latter, is it an A or G?
Ignoring the correction factor, information is defined as the difference between two uncertainties: The above quantity in brackets represents the uncertainty based on the actual nucleotide frequencies, while the value 2 represents the uncertainty if each nucleotide is equally likely to occur. Because there are four possible bases at each nucleotide position, there are between 0 and 2 bits of information at each position: 2 bits if only one nucleotide is present (the quantity in brackets tends to zero as one nucleotide predominates), and 0 bits if each nucleotide is equally likely to occur (the quantity in brackets tends to 2 as each nucleotide becomes equally likely). For example, Figures 3C and 3D illustrate the information values and nucleotide profiles at 20 nucleotide positions flanking the donor sites of our full data set of 10,057 splice sites (based on the Web interface output in Figure 3B).
The Web page example in Figure 3A illustrates that the stored procedure for calculating information and nucleotide profiles has a broad range of options for selecting and analyzing particular subsets of the data set. For example, one can analyze splice sites near introns of a particular length range, or one can compare introns of different types (first vs. internal vs. last intron of a multi-intron gene). Figure 3B shows the results obtained by executing the stored procedure in Figure 3A. By examining different subsets of the data set, one can investigate whether varying a parameter such as intron length might influence the function of the molecular machine that carries out splicing. For example, in our analysis (Weir and Rice, 2004), we found that the amount of information (the level of sequence conservation) is higher at splice sites near larger introns when compared to smaller introns—presumably reflecting the need for stronger interactions of spliceosome components with splice sites flanking longer introns. This analysis of intron length can be carried out using an approach that we call“ progressive partitioning.” By using the stored procedure and systematically increasing the intron length ranges in discrete increments (64-127, 128-255, etc.), one can display the information levels in each range. With progressively longer introns, one finds that the sequence constraints required for successful splicing become increasingly stringent, indicating that the spliceosome system comes under increasing strain. By straining the spliceosome system in this way, more extensive consensus sequences can be defined. In addition to the splice sites immediately flanking the intron whose length is varied, our analysis (Weir and Rice, 2004) indicated that changes in information content are also observed at more distant splice sites, indicating that the spliceosome machinery may participate in long-range molecular interactions—a model that can be tested experimentally.
The analytical approach presented above provides an example of how the relational database encourages one to think informatically about properties of the splicing process by partitioning sequence space. One can imagine extending the partitioning approach by selecting subsets of splice sites with different degrees of mismatch to the consensus sequences and examining compensating changes elsewhere in the splice sites. Indeed, as discussed in Weir and Rice (2004), this approach reveals that poorer matches to consensus are associated with enhanced A content and, to a lesser degree, U content in the vicinity of the splice site. This may facilitate splicing by reducing RNA secondary structure or by increasing association of spliceosome components. One can also imagine analyzing sequence space using different measures such as di-nucleotide or trinucleotide content, or by assessing dependencies with measures such as mutual information (a measure of covariation, in this case between pairs of nucleotide positions). We have begun designing stored procedures to allow the implementation of these and other analytical approaches.
STUDENT EXPERIENCES
We used the Drosophila splice-site database as the basis for a Web-based teaching module in our team-taught bioinformatics course that we offered as an upper-level-undergraduate and graduate-level course in Spring 2003 ( http://mweir.web.wesleyan.edu/igs350/Drosophila_splice_sites.htm). Our class, which had 13 enrolled students (with backgrounds in biology or computer science), was able to work successfully with the database, using two database servers. Making use of stored procedures (see Bioinformatic Analysis without Programming), the students employed the progressive partitioning approach discussed in the section to study the effects of intron length. They also explored the importance of genomic data set quality as it related to the scanning algorithm we used to compute splices sites stored in the database (see Assessing Data Quality and Algorithm Design). Many of our students used the Drosophila splice-site database as a framework for their open-ended term projects (section on Student Term Projects). They used the stored procedures (see Bioinformatic Analysis without Programming) and the structured query language (SQL) query Web interface (see Designing Queries). We expect future term projects of the students to explore some of the new directions discussed at the end of the section on Development of a Genomic Database. Student experiences in the teaching module and term projects provided a rich basis for in-class discussions of how the informatic analysis of splice sites can provide insights into possible molecular mechanisms of the splicing machinery (see Development of a Genomic Database).
Bioinformatic Analysis without Programming
By providing stored procedures for execution in a Web interface, students can often carry out data analyses without actually needing to write programs. In particular, by designing a set of versatile stored procedures for analyzing splice-site data, we have provided our students with a rich environment for analyzing a large genomic data set. The key to our approach is the design of stored procedures with multiple parameters, each of which is relevant for analyzing splice-site data (e.g., minimum and maximum intron lengths, types of introns, or selected tables of genes). These parameters provide students with a built-in versatility by allowing either a simple analysis based on varying one parameter or exploiting the combinatorial power by simultaneously varying several parameters in a data analysis (see Appendix B for a listing of stored procedures).
Figure 3. Computing splice-site information. Web interface (A) shows the parameter options for the stored procedure user_computesplice-siteinfo, which is described in more detail at the link “click here for a description.” Parameter options include, for example, which data sets to use (cDNA Table, Intron Table, Splice-Site Table), which sets of genes to be analyzed (Gene Include Table—none selected in this example) or excluded from analysis (Gene Exclude Table), whether to exclude genes with cDNAs indicating alternative splicing (Alternative Splicing), andother illustrated parameter options. The sample Web page output (B) summarizes the user's parameter choices and shows the result of running the stored procedure, in this case, using donor positions -10 to +10 (where the donor is between -1 and 1) for 10,057 donor sites in 3,090 transcripts. For each nucleotide position, nA refers to the number of donor sites with A at that position (out of 10,057), and pA gives the corresponding proportion. Graphs generated with Microsoft Excel of information (C) and nucleotide content (D) are shown for the output in panel (B).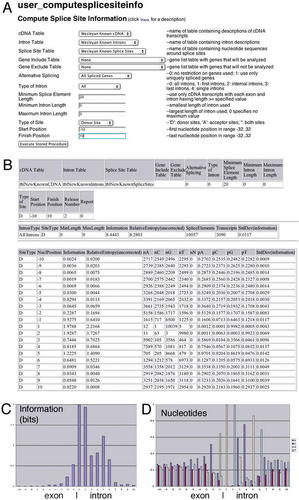
Designing Queries
The broad range of attributes represented by the fields in the database tables provided many opportunities for quite open-ended analysis by students. In addition to the set of stored procedures, we also provided students with a Web-based window in which they can directly enter SQL queries. The SQL language is quite transparent and intuitive, and even with rather simple queries, it is possible to extract interesting data relationships from the database. By studying the primary database schema (Figure 1, Appendix B) and a few examples of queries, it was possible for students to make quick progress.
For example, the query returns the identifiers, ranks, and lengths of all exons between 2,900 and 3,000 bases in length listed in ascending order by length (Figure 4).
select id, rank, finishposition - startposition + 1 as length
from tblNewKnownExons
where finishposition - startposition + 1 between 2900 and 3000
order by length
Testing queries using the SQL query Web interface provides an exciting opportunity for students to explore the splice-site sequence space. They can ask a broad range of questions (e.g., whether exon lengths correlate with information content at splice sites), or they can ask whether there are relationships between general nucleotide content and either intron or exon length. The possible questions are almost limitless.
The SQL query window was used by students in term projects. In our next offering of the course, we will use the query window in a new computer lab module so that all students in the class are introduced to the versatility of the database for studying relationships within biological data. Relationships between attributes of the data set can be assessed through the“ joining” of tables within the database—a process that illuminates all possible associations between components of two tables that satisfy given constraints. Unlike a single flat-file spreadsheet, which can be difficult to navigate for more than one component (gene) at a time, the versatility and flexibility of being able to combine multiple nonredundant tables in many different ways allows one to ask many different kinds of questions about relationships within the data. For example, through appropriate joining (cross-referencing) of tables, one can assess across the whole data set whether poor matches to splice-site consensus sequences are associated with general nucleotide content trends (see Development of a Genomic Database). It is this transparent analytical power of relational databases that allows complex biological relationships to be uncovered.
Assessing Data Quality and Algorithm Design
The Web teaching module in our bioinformatics course ( http://mweir.web.wesleyan.edu/igs350/Drosophila_splice_sites.htm) introduced students to the database and focused on the quality of data resulting from our computation of splice sites. Data quality is a crucial issue in large data sets, as it constrains the potential quality of analysis. The transparency of a well-designed genomic database facilitates the assessment of data quality. Indeed, the SQL language makes it easy to apply constraints to extract a higher-quality data set that is a subset of the full data set. For example, the quality of the data in our splice-site database depended on our algorithm for computing splice sites. As a result of nucleotide polymorphisms (between cDNA and genomic sequences), our algorithm incorrectly predicted some very short introns and exons. Indeed, in the teaching module, students discover that limiting their analysis to cDNAs with introns and exons over 20 nucleotides in length significantly improves the quality of their data set, as measured by higher conformity to the two-base consensus sequences found at each end of the intron. The teaching module introduces students to assessment of the quality of the splice-site-computation algorithm and to considerations of ways to improve the design of the algorithm by taking into account biological context (e.g., the handling of polymorphisms).
Our experience emphasizes the importance of introducing biology students to thinking algorithmically—using precise notation to describe a step-by-step process—thereby encouraging students to think systematically about biological analyses. This can apply to algorithms used in analysis of biological data (e.g., the splice-site computation algorithm described above) or to algorithms developed to model biological machines. Encouraging students to design algorithms that describe the actions of biological mechanisms (e.g., translation by a ribosome) helps the students to build a bridge between bioinformatics and experimental biology and to think about the informatic as well as structural foundations for the biological functions they are studying. Designing algorithms and observing their behavior can reveal steps in a process that were not apparent using apparently systematic thinking. In a different part of our course, we asked students to describe algorithms for biological processes based on finite state machines.
Student Term Projects
In our bioinformatics course, small groups of students, typically including both life-science majors and computer-science majors, worked together on term projects. Their projects included using the Drosophila splice-site database to address questions related to splicing (see Figure 2).
Some of these questions could be addressed using the available stored procedures, and others required some SQL programming. The questions encouraged students to think about different ways to partition the data set and to test for patterns in the data (sequence space) that might reflect constraints on the spliceosome machine. When students use the stored procedures or when they design their own queries in the Web interface, we encourage them to refer to the database schema (Figure 1; Appendix B). By understanding the data structure implied by the schema, students have a framework for considering how relationships within the data can be extracted. This encourages them to consider how different schema frameworks and table designs facilitate asking different kinds of questions. For example, are the database tables well designed, so that it is straightforward to extract the relevant data for analysis (e.g., to analyze information content of aligned sequences or dinucleotide content)? This exposure to database structure encourages students to think about formulating different representations of biological data to answer different questions. In future offerings of the course, we plan to use the database schema as an explicit tool in the lecture component of the course for introducing students to the importance of data representation decisions.
Figure 4. Structured query language Web interface and result of executing query. A sample structured query language query and the results of its execution; the results list all exons between 2,900 and 3,000 nucleotides in length.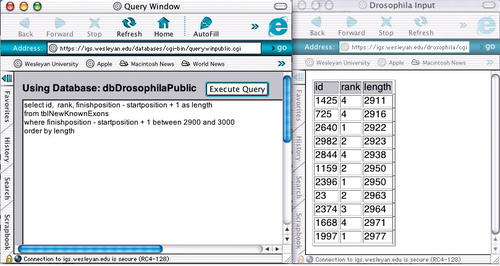
ASSESSMENT
We used our splice-site database in our bioinformatics course in Spring 2003. The course was taught collaboratively by two of the authors: Weir, with a background in biology, and Rice, with a background in computer science. Both instructors attended and contributed to all sessions of the class, and we found the resulting interplay between disciplines to be an extremely effective way to build bridges between biology and computer science. Twelve students enrolled in the course, including the second author (Gladstone), and an additional student audited the class. Standard Wesleyan University teaching evaluations were submitted anonymously by 11 of the 13 students for both Weir and Rice. Of the 22 evaluations, 100% rated the course and teaching in the top two of four categories (Excellent, Good, Fair, or Poor). In the evaluations, students commented on how well they thought the collaborative team teaching worked.
After the end of the course, students were invited to answer an anonymous Web-based questionnaire concerning the splice-site computer lab session. Questions were designed to assess our goals in using databases in teaching, including facilitating students' thinking informatically about large-scale data sets and data set quality, understanding the concept of information, and understanding the algorithm used to compute splice sites. Responses were obtained from nine students (not including Gladstone) and are summarized in Figure 5 and its legend.
In the term projects, several of the student groups used the splice-site database. We noted that compared to the previous offering of the course, when the database was not available, our students this year were able to undertake more sophisticated information-theoretic projects, using the large splice-site data set.
CONCLUSIONS
We are developing several genomic-scale databases to support the information-theoretic analyses of consensus sequences. The generic design of these databases and the systematic use of stored procedures with multiple parameters make the contents of the databases readily understandable and available for analysis. The systematic use of special metadata tables allows the design of a generic Web interface that makes the databases readily accessible to students in courses and also permits the automated posting of new stored procedures without requiring any changes to the Web interface. This versatile database provides a powerful framework for encouraging students to think informatically.
ACCESSING MATERIALS
The Web site of the Berkeley Drosophila Genome Project is found at http://www.fruitfly.org. The Drosophila splice-site database described in this paper is found at the Wesleyan Integrative Genomic Sciences Web site, http://igs.wesleyan.edu. A sample Web-based teaching module that makes use of this database is found at http://mweir.web.wesleyan.edu/igs350/Drosophila_splice_sites.htm. The link at http://igs.wesleyan.edu/metatables describes the use of metadata tables for automatic Web posting of stored procedures.
Figure 5. Assessment: This Web-based questionnaire was completed by 9 of the 13 students in the class. Each assessment scale (1 to 5) with descriptors is shaded; numbers of student responses are in bold. The survey indicated that all students found the database useful for analyzing large-scale data sets (question 2a). Students appreciated having practical examples of data sets to work with, rather than just working with abstract notions (2b, 4b). Most students felt they did not need substantial programming experience (3a). Most students found the stored procedures useful (3b, 3c), but several had some difficulty with the Web interface (5a) and suggested help guides explaining the procedures (5b). Many felt that the lab session helped them understand the algorithm used to compute the splice sites (3d), and most felt informed about assessing data quality (3e), which was one of the goals of our lab. Most thought the lab helped them to understand information content (4a) and to think informatically about biological problems (6a). One student appreciated introducing the concept of “algorithm” when thinking about biological processes (6b). All but one student felt that the relational database setting facilitated their understanding of concepts (7), and many thought relational databases would be useful for other biological data sets (8). Samples of student responses to the written-response questions include 2b It is fast, capable of analyzing huge sets of data, easy to use. Concern regarding understanding stored procedures. 2c Sometimes it was hard to conceptualize where the data was coming from. 4b Practical examples illustrate the concepts presented in class in a manner that is easier to grasp than mere abstractions. It helped to understand the complexities of understanding how to utilize the information. 5b If you can include a very detailed HELP or GUIDE web page and explain thoroughly how to use the interface, it will make it easier. [Several students expressed this sentiment.] 6b Thinking [about a] biology problem in a mathematical way is very new and very interesting. I learned that algorithm[s] [are] really important in understanding a biological problem in a quantitative way.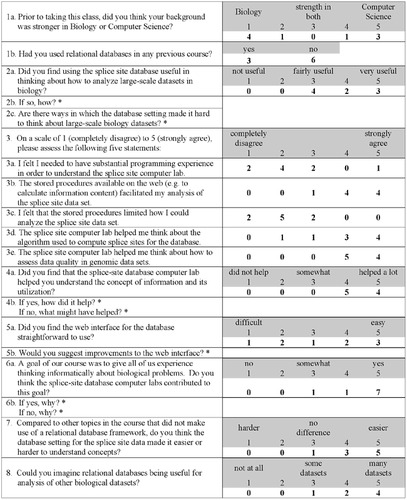
APPENDIX A RELATIONAL DATABASES
Modern relational database management systems provide a powerful framework for representing and manipulating large-scale genomic data. Their utility rests on several basic principles.
The standard representational framework of a relational database (O'Neil and O'Neil, 1999) is ideally suited for organizing and representing genomic data. Different types of properties of genes (such as functional annotation and sequence data) can be assigned to different tables in the database, and in each table, different attributes of the property (such as gene name, associated protein, and transcription start position) can be stored. We can think about the genome as a whole being represented by the database, where each gene property is represented by specific values of standardized attributes, rather than trying to think about thousands of genes with unorganized, separate characteristics.
The power of the relational database structure is that organizing complex genomic-scale data in this framework exposes the data set to uncovering relationships between properties of the data. The structured query language (SQL) of relational databases provides the ability to answer straightforward questions about the data without needing to write extensive code defining search algorithms, because the organization naturally leads to common analytical approaches which can be expressed in the SQL language. A relational database allows you to perform a task such as, “list all Drosophila genes that have a transcription start site less than 100 KB from the end of the left arm on chromosome 3” by executing an SQL query of the form
select genename
from tableGenes
where transcriptionstart, 100000 and arm = '3L'.
Moreover, sophisticated queries can often be written in a simple procedural language supported by the database that extends standard SQL. A standard approach is to use this language to write stored procedures that permit a more in-depth data analysis by allowing many different combinations of parameter values to be tested.
A stored procedure that summarizes the preceding query might have the name ListGenes and parameters corresponding to the arm and distance from the start of the arm, but it might also have a parameter that restricts the types of genes that will be listed. For example, the following code specifies the execution of ListGenes and lists all ribosomal genes that have a transcription start site less than 50 KB from the end of arm 2R:
exec ListGenes 50000, '2R', 'ribosomal'.
In summary, relational databases allow users to formulate and answer basic questions about the data and allow database developers to create more sophisticated procedures for analyzing the data. In fact, the design of queries and stored procedures is an ongoing process—the results of a given analysis lead to the need to develop new and progressively more sophisticated stored procedures. In our case, we have developed an approach for recording new stored procedures so they are automatically made available to students through a Web interface to the database (see Appendix B).
APPENDIX B DESIGNING THE SPLICE-SITE DATABASE
The main issue in database design is the selection of entities to represent as tables and the choice of attributes of these entities to represent as fields in the tables. Of course, the particular type of genomic data that is being represented will determine the specific types of tables and fields that will be needed. In our case, to design a splice-site database, we decided to represent four key entities: cDNA-gene correspondence, exons, introns, and splice-site regions. On the basis of these entities, one can define a set of attributes to facilitate storing the results computed by the algorithm for the splice-site specification. For example, the cDNA-gene table has fields for storing cDNA transcript identifiers, the corresponding gene identifier, and the name of the gene. The intron table has fields for storing the start and finish positions of each intron on the genomic DNA and fields for storing the number of nucleotides of each type present in each intron. The splice-site table has fields for storing the type of splice site (donor or acceptor), the nucleotide position (32 bases upstream or downstream of the splice site), and the specific nucleotide found at each position.
The four primary tables that are used to store the computed splice-site data are shown in Figure 1. By combining the data in these tables in a variety of ways, complex queries and procedures can be written to analyze virtually any aspect of the splice-site data. For example, the splice-site database currently provides eight specialized procedures with a variety of parameters for analyzing data (listed in Figure 6A). Below, we provide an overview of the design of the procedure user_countpattern that counts the matches to a pattern in splice-site regions. Students in our bioinformatics courses also have the capability of running their own custom queries in the database by using a special structured query language query window in the Web interface (see Designing Queries).
As described above, the primary data tables store the computed splice-site data. These tables support the stored procedures that are available for analyzing the data as well as ad hoc user queries. The schema for the data tables is shown in Figure 1. Each row in each table diagram represents a separate field. For example, the splice-site table tblNewKnownSpliceSites contains the fields sitetype (indicating a donor or acceptor site), nuclposition (an integer between 32 and +32 [excluding 0] that denotes a position upstream [-] or downstream [+] of the splice site), and nucleotide (one of the four possible nucleotides A, C, G, or T).
Figure 6. Web pages for stored procedures. (A) The web page for choosing a stored procedure. A set of eight stored procedures is currently available. (B) The Web page for the stored procedure user_countpattern showing the parameters that can be defined by the user, including the pattern to be searched and constraints on intron lengths and nucleotide positions. The sample parameters would specify searching for and counting cases of“ GGT[AG]AGT” between donor site positions -10 to 10 for introns of length ≤1,000 in cDNAs where all introns and exons have length ≥20.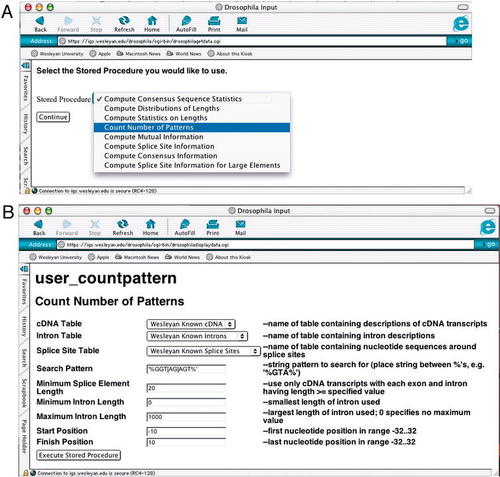
In each table diagram, the set of gold keys represents the primary key. This is a collection of fields that uniquely determine the values of the other fields in the table. For example, in the intron table tblNewKnownIntrons, the primary key consists of the fields id, rank, and release. In other words, each intron entry is determined by an integer identifier, the rank of the intron (1, first; 2, second; etc.), and the release number of the data set from the Berkeley Drosophila Genome Project ( http://www.fruitfly.org) that was used to compute the splice-site data. Similarly, the fields id and release represent the primary key of the table tblNewKnowncDNA. This pair uniquely determines the cDNA-gene correspondence represented by the cDNA transcript ctstring and the gene cgstring.
The lines between table diagrams indicate relationships that are called referential integrity constraints. These constraints enforce data dependencies between certain pairs of tables. For example, the line between the tables tblNewKnowncDNA and tblNewKnownExons indicates that every value of id in the second table must also be present in the first table. Similarly, every triple of values id, intronrank, release found in tblNewKnownSpliceSites must be present as values of id, rank, and release in tblNewKnownIntrons.
Figure 7. Results of executing user_computepattern. The illustrated output indicates that most cases of the search pattern “GGT[AG]AGT” are found at donor nucleotide position -1.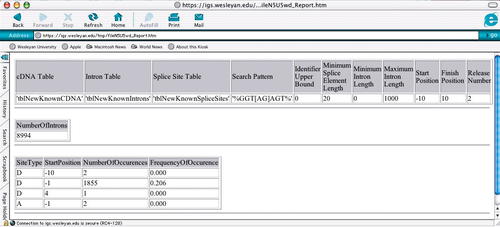
Each stored procedure uses a combination of the data entries in the tables to compute its result set. For example, Figure 6A shows the Web interface page used to select a particular stored procedure. Figure 6B shows the Web page that is generated by selecting the option “Count Number of Patterns” that corresponds to the stored procedure user_countpattern. By selecting three of the four tables shown in Figure 1 and specifying the contents of the remaining textboxes (Figure 6B), one can use this Web page to execute user_countpattern with the desired parameters. Figure 6B illustrates parameters for execution of user_computepattern, using the donor splice-site consensus sequence “GGT[AG]AGT,” restricting attention to the cDNA transcripts where every exon and intron contains at least 20 nucleotides and the introns contain at most 1,000 nucleotides.
The stored procedure computes the number of exact matches to the consensus sequence in the restricted data set as well as the starting positions of the matches in the range from -10 to 10. The results of executing the procedure are shown in Figure 7. As expected, the majority of the matches (1,855) are found starting at the donor position -1. In these cases, a G is found at position -1, a G is found at position 1, a T is found at position 2, either an A or a G is found at position 3, and so on.
The manner in which the actual computation is performed by the procedure is outlined here: First, the constraint on the minimum splice length (20) is used to create a new temporary table containing the entries in the cDNA table that satisfy the constraint. Second, the entries in the temporary table are joined with the entries in the intron table that have matching ids and release numbers and satisfy the minimum and maximum length constraints 0 and 1,000, respectively, to create a second temporary table. Third, the entries in the second temporary table are joined with the entries in the splice-site table that have matching ids, ranks, and release numbers to create a temporary splice-site table. Finally, the entries in the temporary splice-site table are used to generate the nucleotide sequences found at the positions 10 to 10 for both donor and acceptor sites. The sequences that match the pattern“ GGT[AG]AGT,” starting at some position in the range from -10 to 10 are computed using a built-in pattern matching routine. The starting positions and the numbers of these sequences are displayed in Figure 7.
The database also contains a collection of metadata tables. In general, the term metadata refers to the fact that the tables contain information about genomic data and procedures as opposed to actually containing the genomic data. In the splice-site database, the metadata tables store a variety of information about the stored procedures. For example, there are tables that contain the names of the stored procedures displayed in the Web interface, the names of the parameters for these procedures, and lists of possible choices for the actual values of certain parameters. There is also a metadata table that contains the names of other tables that store lists of specialized genes (e.g., the genes on a particular chromosome arm or the genes used in a particular experiment).
The metadata tables support several specialized views (dynamically generated tables) and procedures that compute summary information. A scripting program uses the summary information from the metadata to generate the layout of the Web pages that provide an environment for executing the stored procedures. This approach allows the script to be written in a generic manner independent of specific stored procedures and permits additional stored procedures to be added to the database and exported for use without making any changes to the code. Details of this approach are described at http://igs.wesleyan.edu/metatables.
FOOTNOTES
Monitoring Editor: Raquell Holmes
ACKNOWLEDGMENTS
We thank Robert Lane, Laurel Appel, and Sylvia Weir for useful discussions. We also thank two anonymous reviewers for their suggestions. This work was supported in part by funds from the Howard Hughes Medical Institute Undergraduate Science Education Program to Wesleyan University, and by a grant from the W. M. Keck Foundation.



