Computer Simulations Improve University Instructional Laboratories
Abstract
Laboratory classes are commonplace and essential in biology departments but can sometimes be cumbersome, unreliable, and a drain on time and resources. As university intakes increase, pressure on budgets and staff time can often lead to reduction in practical class provision. Frequently, the ability to use laboratory equipment, mix solutions, and manipulate test animals are essential learning outcomes, and “wet” laboratory classes are thus appropriate. In others, however, interpretation and manipulation of the data are the primary learning outcomes, and here, computer-based simulations can provide a cheaper, easier, and less time- and labor-intensive alternative. We report the evaluation of two computer-based simulations of practical exercises: the first in chromosome analysis, the second in bioinformatics. Simulations can provide significant time savings to students (by a factor of four in our first case study) without affecting learning, as measured by performance in assessment. Moreover, under certain circumstances, performance can be improved by the use of simulations (by 7% in our second case study). We concluded that the introduction of these simulations can significantly enhance student learning where consideration of the learning outcomes indicates that it might be appropriate. In addition, they can offer significant benefits to teaching staff.
INTRODUCTION
Laboratory classes are a feature of most degree programs in university biological science departments. Indeed they are considered an essential part of most biological degrees, particularly for those students wanting to continue their careers in science. They can, however, be cumbersome and unreliable in producing the required data that, in turn, places a drain on staff time and resources. Coupled with the increasing number of students admitted to universities, this can put a great amount of pressure on department budgets. Computer-based simulations of student practical classes (“virtual laboratories”) can provide a cheaper and time-saving alternative to traditional practical classes. Indeed, several studies have considered the simulation of a range of skills and practical concepts in the biological sciences (Dewhurst and Williams, 1993; Dewhurst et al., 1992; Hughes, 2000; Maury and Gascuel, 1999; Modell, 1989). These studies highlight many suggested benefits that are common to most types of computer-based learning (CBL) and include:
Flexibility of time (students can complete the virtual laboratory at a time convenient to them; Race, 1994).
Flexibility of location (students can complete the virtual laboratory in a location other than the teaching laboratory; Race, 1994).
Control of learning pace (students can take as long as required to understand the concepts with the virtual laboratory). Heerman and Fuhrmann (2000) highlight this in the context of students not having to rush to vacate the laboratory; hence, they can work at their own pace.
It has also been suggested that virtual laboratories offer an additional set of potential advantages as follows:
Decreased marking time (simulations can be developed to perform the marking through computer-based assessment and then send marks directly to the tutor electronically; O'Hare et al., 1998).
Decreased laboratory time (students do not need to be in the potentially dangerous laboratory environment; Dewhurst and Williams, 1993; Dewhurst et al., 1992).
Decreased infrastructure costs (it is not necessary to spend funds on laboratory equipment, reagents, consumables, and laboratory hire). Virtual laboratories have been seen to significantly cut the costs of running a practical class (Dewhurst et al., 1994; Leathard and Dewhurst, 1995).
Research in this field also challenges the notion that computer-based simulations are, in some way, inferior to “real” practical classes, suggesting that student performance in assessments is comparable (Dewhurst et al., 1994; Leathard and Dewhurst, 1995). In this regard, Hughes (2000) adds the caveat that this is only appropriate if the learning outcomes of the practical class do not include the development of laboratory-based skills. The ability to use laboratory equipment and reagents or manipulate test animals can be an essential learning outcome of a practical class, but this is not always the case. For instance, if the learning outcomes focus on the interpretation and manipulation of data, virtual laboratories can provide workable alternatives.
In this study, we test the hypothesis that, in certain situations, computer simulations can provide an improvement in student learning compared with real, or traditional, laboratory classes. For the purposes of this study, improvement is measured objectively either as a decrease in the time taken for students to study to a given level of performance (efficiency) or by an increase in the marks they achieve in assessment (effectiveness). To test this hypothesis, we evaluated two simulations, the first in chromosome analysis (karyotyping) and the second in bioinformatics.
In all cells, chromosome condensation occurs in a very ordered fashion, and the distinct pattern of chromosomes (karyotype) is easily recognizable to the trained eye for most organisms. The ability to karyotype humans is essential in clinical diagnostics and physical gene mapping and is thus a skill taught in practical classes in many university biological science departments. Conversations with colleagues reveal that, traditionally, students are given a photograph of a chromosome preparation, scissors, and glue and asked to cut out, arrange, and stick the chromosomes in the correct order (Paris Conference, 1971). It has been suggested to us through several personal communications that large amounts of time are spent cutting and pasting, leaving proportionally less time available for analysis; this is also supported by our own experience. Other colleagues have, however, suggested that the physical cutting and pasting does not significantly affect time and, in any event, is a task that is enjoyed by the students. With this in mind, we propose the hypothesis that a computer-based simulation provides a significantly quicker alternative to cutting and pasting and leads to higher marks in the subsequent assessment.
The second study involves a practical class in bioinformatics (genome analysis). Skills that students need to develop include accessing existing Human Genome Mapping Project databases and answering a variety of biological questions directly at the computer terminal. Traditionally these classes are taught by didactic lectures and practical computer laboratories. A tutor would take the class through each stage in selected examples. In this study, we test the hypothesis that students learn more effectively in a bioinformatics class that involves a set of computer-based lectures and computer simulations of database navigation compared with the traditional approach.
METHOD
We report the results of two studies. Both studies involved the development of computer-based simulations: one to teach the skill of karyotyping and one to teach the principles of introductory bioinformatics. Both of the simulations were based around the Virtual Lecture interface proposed by Evans and Edwards (1999) with the use of Macromedia Authorware 6.0 and additional interactive programming added where necessary. Among the special usability features of the interface is the requirement that material is divided into a set of topics and subtopics designed to allow ease of navigation. Each subtopic consists of series of pages that can be read in a linear fashion.
Study 1: Chromosome Analysis
In the first study, a cohort of level-one undergraduates in the Department of Biological Sciences at Brunel University (West London) were recruited. To match the groups as much as possible according to academic ability, students were pretested on their knowledge of basic genetics, and this data was used to divide them into two groups of equal size and ability. Group A undertook the traditional approach to learning the skill of chromosome analysis involving a photograph, scissors, and glue (Figure 1), whereas group B undertook the computer-based simulation approach. The development of the aforementioned computer simulation (“KaryoLab”) has been reported by us previously (Gibbons et al., 2003) and involves a series of drag and drop interactions that were incorporated into the program with images of individual chromosomes (Figure 2). Both groups were given a 30-minute lecture on the rudiments of karyotyping and one karyotyping exercise to perform in order to learn and practice the skill of analysis. For both groups, the chromosome images were identical. Group A received formative feedback from their tutor on their performance, whereas in group B in the virtual laboratory, if chromosomes were positioned incorrectly, they returned immediately to their original position. In a second exercise, both groups were given an identical image of a chromosome preparation. Group A did the exercise with scissors and glue and group B with KaryoLab. In this case no formative feedback was given and student performance was assessed. The time involved in completing the learning section and the assessment was not constrained for either group; however, location was constrained to prevent collusion between groups. The time taken for each student to perform both sections was logged so comparisons could be drawn. For the virtual laboratory (KaryoLab), marking was automatic and a feature of the Authorware 6.0 software. For the students undertaking the traditional approach, marking was performed by a tutor. Any differences between the marks for groups A and B were evaluated by an unpaired one-tailed Student's t-test.
Figure 1. Image of student cutting and pasting chromosomes.
Figure 2. Example of chromosome analysis in KaryoLab.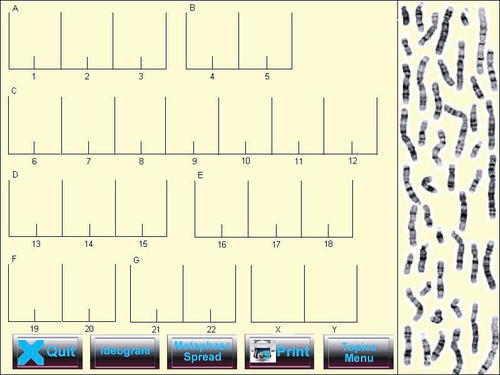
In addition, student opinion of KaryoLab was evaluated by 10 final-year undergraduates in the Biological Sciences Department. These were a different subset of students than did the comparative studies outlined above who did both exercises. Students were asked 18 closed questions on a five-point Likert scale.
Study 2: Bioinformatics
The virtual laboratory for bioinformatics was written in the same learning environment as KaryoLab. In this case the virtual laboratory consisted of“ virtual lectures” (Evans and Fan, 2002) on the subject material (which included the use of NCBI and NIX databases), followed by an exercise in genome database searching (see Figures 3, 4). The exercise involved a simulation of the appropriate databases with relevant instruction of how to perform a series of example operations (e.g., searching for the alkaline phosphatase gene). This compares with the traditional approach in which students are given an oral lecture by the tutor and then asked to perform the same exercise on the real databases overseen by the tutor. Specifically, students were given instructions on how to perform the various analyses and how to identify CpG islands, definitions of specific terms such as STS and clones, and the use of different NCBI databases such as Map Viewer, Locus Link, and Unigene. In both “real” and “virtual” modes, the subject matter was the same. The use of a simulation of a database rather than a real database has the advantage that it is possible to trap mistakes made by the student before the consequences have a drastic effect on the whole experiment. This is much like the early spotting of a mistake in the use of physical equipment in a real laboratory. The major differences between the lecture-delivered and the electronic-delivered styles are summarized in Table 1. In this study, a cohort of level-two undergraduates in the Department of Biological Sciences at Brunel University were recruited. These students were randomly assigned into one of two test groups (A and B). The bioinformatics teaching material and exercises were divided into two topics (1 and 2). In this case the time taken was roughly the same for both real and virtual exercises. That is, the students were given 1 h to complete each of the virtual lectures and a 3-h session for each of the practical exercises.
| Real | Virtual | |
|---|---|---|
| Chromosome analysis | Students cut out chromosomes with scissors | Chromosomes are precut and presented on screen |
| Students physically cut out photographs then stick down with glue once completed | Drag and Drop function is used | |
| Students need to wait for tutor to mark exercise | Mark is given immediately | |
| Tutor needs to mark all exercises | Mark sent directly to tutor | |
| Bioinformatics | Tutor gives lectures to cohort | Lecture given to students in electronic format, can be done at own time, place, pace |
| Tutor talks through worked examples trying to ensure all students can keep to same pace | Students are guided through worked examples by tutorial | |
| Tutor and demonstrators give constant feedback to students when they go wrong | Formative feedback given in worked exercise |
The experimental design involved both groups experiencing both teaching techniques (real and virtual). This allows a comparative analysis to be undertaken, obviates the need to pretest the students, and has the advantage that the sample size is effectively doubled. To achieve this, group A undertook topic 1 using the virtual approach and topic 2 with the real approach. Conversely, group B undertook topic 1 with the real approach and topic 2 with the virtual approach. At the end of each topic, both groups were given the same assessed assignment on the material. The assessments were carried out under examination conditions, and both groups were kept separate during the learning process and the assessments to prevent students from different groups communicating. For this study both the time and location were constrained. The assignments were marked anonymously by the tutor in both approaches. We felt that this approach was not appropriate in study 1 because karyotyping is a generic skill applicable to all G-banded chromosome preparations. Thus, students would learn karyotyping by one means in the first exercise and then, if the groups were reversed, use that knowledge in the second, negating the significance of the results comparison.
Any significant differences were evaluated by one-tailed Student's t-tests.
Figure 3. Example of database searching in the bioinformatics virtual laboratory. Here the navigational interface is shown, and in the main screen, the NCBI homepage is displayed. The tutorial is in the process of guiding the user to the “Map viewer” site (red ring).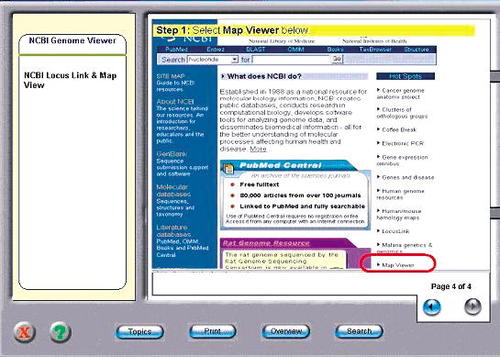
RESULTS
Study 1
Assessment Marks for KaryoLab. A total of 47 students took part in study 1. The results for the assessment marks are displayed in Table 2. Although the marks for group B (using KaryoLab) were about 4% higher, this difference was not statistically significant (unpaired samples t-test, t(45) = 0.68, one-tailed p = .25).
| Group A (real) | Group B (virtual) | |
|---|---|---|
| N | 24 | 23 |
| Mean | 43.2% | 47.6% |
| Standard Deviation | 12.8 | 15 |
Time Analysis. The results of the time analysis are displayed in Table 3. In both cases, practice and assessment, group B using KaryoLab was able to complete the sections faster than group A. In the practice session, group B took almost a quarter of the time (unpaired samples t-test, t(30) = 11.96, one-tailed p, .001). In the assessment itself, group B (using KaryoLab) took less than half the time taken by group A, who all used scissors and glue (unpaired samples t-test, t(32) = 8.03, one-tailed p, .001).
| Group A (real) practice | Group B (virtual) practice | Group A (real) assessment | Group B (virtual) assessment | |
|---|---|---|---|---|
| N | 23 | 24 | 23 | 24 |
| Mean (min) | 77 | 20 | 68.3 | 33.8 |
| Standard deviation | 21.7 | 8.5 | 19.5 | 8.6 |
Questionnaires. The results for study 1 questionnaires are in Table 4. Because these were a different set of students than those who did the tests and because these students did both exercises compared with those above, who only did one, it was not possible to triangulate qualitative and quantitative data in this case. The responses were, however, generally positive. The most notable response was 100% of the students asked would have preferred to complete KaryoLab over the real lab with the scissors and glue method.
| Statement | Agree | Neutral | Disagree |
|---|---|---|---|
| Introduction | |||
| Aims are set out clearly | 100% (10) | 0% (0) | 0% (0) |
| Learning outcomes are pitched at an appropriate level and are clearly set out | 100% (10) | 0% (0) | 0% (0) |
| Assessment guidelines are clear and are easy to understand | 100% (10) | 0% (0) | 0% (0) |
| Background information | |||
| Chromosome information is informative and appropriate to the virtual lab | 100% (10) | 0% (0) | 0% (0) |
| Karyotyping information is informative and appropriate to the virtual lab | 100% (10) | 0% (0) | 0% (0) |
| Aberrations information is informative and appropriate to the virtual lab | 100% (10) | 0% (0) | 0% (0) |
| Right way up? | |||
| The instructions are clear and easy to follow | 100% (10) | 0% (0) | 0% (0) |
| The assessment is easy to complete | 100% (10) | 0% (0) | 0% (0) |
| The assessment provides appropriate feedback | 50% (5) | 30% (3) | 20% (2) |
| Karyotypes | |||
| The instructions are clear and easy to follow | 100% (10) | 0% (0) | 0% (0) |
| The assessments are easy to complete | 100% (10) | 0% (0) | 0% (0) |
| The resolution of the chromosomes is sufficient for karyotyping to be performed easily | 100% (10) | 0% (0) | 0% (0) |
| The diagnosis is easy to make from the information included in the karyotype | 90% (9) | 10% (1) | 0% (0) |
| The assessment provides appropriate feedback | 100% (10) | 0% (0) | 0% (0) |
| Overall | |||
| The interface design is well laid out | 100% (10) | 0% (0) | 0% (0) |
| The format is clear and easy to follow | 100% (10) | 0% (0) | 0% (0) |
| Each section is appropriate to the aims of the lab and should be included in the final program | 90% (9) | 10% (1) | 0% (0) |
| I would have liked the opportunity to complete this virtual lab instead of the more traditional laboratory procedures for learning how to karyotype. | 100% (10) | 0% (0) | 0% (0) |
The tutor also reported that it was much easier to perform practical classes with KaryoLab than with the scissors and glue approach, in that students (after about half an hour of tuition) could go off and do the exercise in their own time. In contrast the scissors and glue approach required a dedicated 3-h session with postgraduate “demonstrator” help and the inevitable hazards of losing cut-out chromosome images because of open windows, passing colleagues, coughing, sneezing, sighing, etc. Now KaryoLab has completely replaced the scissors and glue approach in our classroom because of the perceived increase in popularity.
Study 2
Assessment marks. A total of 30 students took part in study 2. Topic 1 was studied and assessed 1 wk before topic 2. The number of students in each group and their mean marks are given in Table 5. Collapsing the groups (and removing students who didn't take both tests for direct comparisons on the same students) gives a mean score of 59.5% for students doing the virtual lectures and simulation and 58.0% for students receiving a real lecture and traditional laboratory session. However this difference is not statistically significant (paired samples t-test, t(24) = 0.25, one-tailed p = .40). Results are similar if the three students who took topic 2 but not topic 1 are included.
| Topic | Topic 1 | Topic 2 | ||||
|---|---|---|---|---|---|---|
| Group/delivery | Group A (virtual) | Group B (real) | Group A (real) | Group B (virtual) | ||
| N | 13 | 14 | 16 | 14 | ||
| Mean | 53.0% | 45.6% | 69.7% | 59.7% | ||
| Standard deviation | 10.6 | 10.9 | 23.7 | 27.6 | ||
The experimental design allows for a possible interaction between the topic studied and the delivery (real or virtual); therefore, we should also consider the uncollapsed results.
For topic 1, the mean score was 7.4% higher in the virtual mode compared with the real mode. This result is statistically significant (unpaired samples t-test, t(25) = 1.78, one-tailed p = .04).
For topic 2, by contrast, the mean score was 9.9% lower in the virtual mode compared with the real mode. This result is not, however, statistically significant (unpaired samples t-test, t(28) = 1.04, one-tailed p = .15).
In this case, the time to complete the exercises was roughly the same for real and virtual exercises; however, this outcome reflects the fact that time was restricted for these exercises. Anecdotally it seems that the students were taking roughly the same time to finish the exercises, but the range was greater in the virtual exercises, with some students finishing much earlier and others taking more time. The tutor reported that the practical sessions were much less stressful; that is, they were not being called for assistance by quite so many students at one time and not as many postgraduate student“ demonstrators” were needed.
DISCUSSION
The results from study 1 show that our virtual karyotyping laboratory (KaryoLab; Gibbons et al., 2003) can achieve a substantial decrease in study time (by a factor of four) without any significant effect on student performance in assessment. Thus virtual laboratories can serve as remarkably efficient learning mechanisms in this subject area. This conclusion is reinforced by the general preference students showed for the use of a simulation rather than the traditional approach. The radical rationalization of time in this exercise means that students should be able to complete more exercises within the same given time frame and thus, in theory, improve their learning. Study 2 indicates that, in certain circumstances, virtual laboratories can improve the performance of students in assessment (by over 7% for topic 1). Thus virtual laboratories can be significantly more effective learning mechanisms than real ones in this subject area also. This result appears to be, however, dependent on the nature of the material presented because topic 2 showed no significant difference in student marks. The reader will note that the mean marks for topic 1 were substantially lower than for topic 2 (by 15%). One possible interpretation of this is that the subject matter of topic 1 was harder to learn than that of topic 2. This is consistent with the subjective evaluation of the subject material made by most bioinformatics lecturers. This would suggest that the benefits of virtual laboratories are greatest when the level of difficulty of the material is not too low. This is consistent with studies that have indicated that multimedia and computer-based learning are most effective when the media is presented to learners with low prior knowledge or aptitude (Najjar, 1996), although these attribute their results to the knowledge of the students rather than level of the material. Another explanation of course is that the exploration of topic 1 provided the students with the skills that enabled them to perform better on topic 2. Further investigations are necessary to determine the extent of the validity of the interaction between mode of delivery and difficulty of material. For instance a four-way study with easy and hard topics, real versus virtual delivery, and outcomes measured by tests and opinion questionnaires similar to this study might go some way toward addressing this question.
The use of computer-based simulations in undergraduate practical classes is becoming more widespread (Dewhurst et al., 1992, 1994; Hughes, 2000; Maury and Gascuel, 1999; Modell, 1989). For the most part however, studies concentrate on making the experience at least as good or nearly as good as the real practical class. The benefits of safety and flexibility are often highlighted in these studies. To the best of our knowledge however, this study is among the first to consider the ability of a computer-based simulation to achieve a significant improvement in student learning. Clearly in instances where students are required to manipulate laboratory equipment or reagents, computer simulations would not be appropriate. In the two cases considered in this study—chromosome analysis and bioinformatics—the use of a virtual laboratory can represent an overall improvement. The issue of when and whether computer-based simulations should be considered for simulating practical classes is entirely dependent on the intended learning outcomes. In study 1 (karyotyping) the intended learning outcome was that students should be able to accurately analyze chromosomes. For the real laboratory, the necessary skill of accurately cutting and pasting chromosomes from a photograph was not one of the intended outcomes. Thus, achievement was measured by their karyotype mark and, although these were not significantly different for each group, the group using KaryoLab were able to complete the exercises in a much shorter time frame, thereby allowing them to practice more karyotypes (and hence hone their skills) more efficiently. In study 2 (bioinformatics) the learning outcomes were the accurate and confident use of the NIX and NCBI databases. These learning outcomes were measured by the tests described in this paper. In this case the use of computer simulation is not only entirely consistent with the intended outcomes, but also helps to reinforce them.
This paper investigates the effect of short-term learning but does not address the issue of whether learning practical exercises via multimedia reinforces long-term student learning compared with traditional approaches. Previous research, however, suggests that computer-based packages show a significant improvement in both the short term and long term for deep learning, as shown with transfer tests (Mayer et al., 2003). To the best of our knowledge, the effect of multimedia-based approaches on long-term learning has yet to be tested in the context of simulations of student practical classes, and this will form the basis of future studies in our group.
Finally it is important to note that, although the main advantages of the use of virtual laboratories are for the students and their learning, there are also important benefits for lecturers. The time spent marking assessments can be almost eliminated by integrated computer assessment, and the time spent lecturing can be considerably reduced by the provision of virtual lectures.
We suggest that the results presented in this study provide evidence of the advantages of computer-based practical classes over traditional ones, at least in the subject areas presented. Combined with the advantages they offer in terms of flexibility in time, location, pace, and process, they can offer a potentially more efficient mode of teaching for lecturers and a more effective and efficient mode of learning for students. Further studies will establish examples of other practical class scenarios to which this pertains.
FOOTNOTES
Monitoring Editor: Eric Chudler
1 Potential conflict of interest: The authors were involved in both production and review of the products reported in this paper.
ACKNOWLEDGMENTS
The work for this paper was partially supported by a grant from the Learning and Teaching Development Unit at Brunel University and partially by the Brunel Enterprise Centre. The work presented here was carried out under the auspices of the Virtual University Project at Brunel University. We are grateful to the staff of the clinical cytogenetics unit, University of Newcastle, for providing the chromosome images.



