A Combination of Hand-held Models and Computer Imaging Programs Helps Students Answer Oral Questions about Molecular Structure and Function: A Controlled Investigation of Student Learning
Abstract
We conducted a controlled investigation to examine whether a combination of computer imagery and tactile tools helps introductory cell biology laboratory undergraduate students better learn about protein structure/function relationships as compared with computer imagery alone. In all five laboratory sections, students used the molecular imaging program, Protein Explorer (PE). In the three experimental sections, three-dimensional physical models were made available to the students, in addition to PE. Student learning was assessed via oral and written research summaries and videotaped interviews. Differences between the experimental and control group students were not found in our typical course assessments such as research papers, but rather were revealed during one-on-one interviews with students at the end of the semester. A subset of students in the experimental group produced superior answers to some higher-order interview questions as compared with students in the control group. During the interview, students in both groups preferred to use either the hand-held models alone or in combination with the PE imaging program. Students typically did not use any tools when answering knowledge (lower-level thinking) questions, but when challenged with higher-level thinking questions, students in both the control and experimental groups elected to use the models.
INTRODUCTION
It is essential for life science students to understand that biological structure determines function across all size scales. Molecular biology instructors commonly list the understanding of protein structure/function relationships as a learning objective for their students, because students who understand these relationships can link genetics with biochemical pathways and higher processes. But how do we help our students understand structures they cannot see or touch? At the macroscopic level, vertebrate morphologists study skeletal muscle attachment positions on bones to determine how much force muscles exert. Students draw upon their visual and tactile experience with macrostructures and arrange new, more intellectually challenging concepts on this pre-existing framework. For example, students find it easier to understand how skeletal muscle insertion points affect joint openings and closures after reflecting on (and testing out) how their own bodies work. At the microscopic level, cellular biologists examine cell membrane receptor structure to identify key amino acid residues involved in binding a ligand to initiate a signal transduction pathway. It is not as obvious, however, for a student to grasp spatially and abstractly the impact of amino acid sequence on secondary, tertiary, and quaternary structure, which in turn affects protein function. In both of these examples, researchers and students gain a more thorough and accurate understanding of their system after they comprehend how a structural feature affects function.
Molecular biology instructors have traditionally used two-dimensional imagery to represent microstructures to students. To examine microstructures in the laboratory, the best tool that most public school and undergraduate biology classrooms offer is light microscopy, typically magnifying structures 100 to 1000× actual size. This relatively low magnification means that subcellular structures smaller than mitochondria remain invisible. Accurate examination of molecular structure requires the use of x-ray crystallography or nuclear magnetic resonance (NMR) technologies to reveal structural details on an atom-by-atom basis. Fortunately, the results of x-ray crystallography or NMR studies are freely available through the protein data bank (www.pdb.org) (Berman et al., 2000) and can be used to generate three-dimensionally accurate images using web-based molecular imaging programs such as MDL Chime, Jmol (an open-source Java viewer for chemical structures in 3D [www.jmol.org]), and Protein Explorer (PE; Martz, 2002). Bioinformatics tools using websites such as that administrated by the National Center for Biotechnology Information have increasingly been used as teaching tools.
Two-dimensional pictures of molecules based on crystallography and NMR data are becoming more common in our biochemistry and introductory biology books, but many of these textbooks predominantly use multicolored “blobs” to indicate protein, nucleic acid, and carbohydrate molecules (see Figure 1A). Authors typically use simplistic imagery when their goal is to emphasize larger concepts such as how multiple phosphorylation events in a chain of molecules result in signal transduction. Although these images provide a broad overview, this type of imagery does not allow students to fully appreciate the precise, often subtle, molecular conformational changes that ultimately allow cells to sense and respond to environmental changes. At worst, simplistic imagery could introduce misconceptions to students, particularly those new to learning molecular biology.
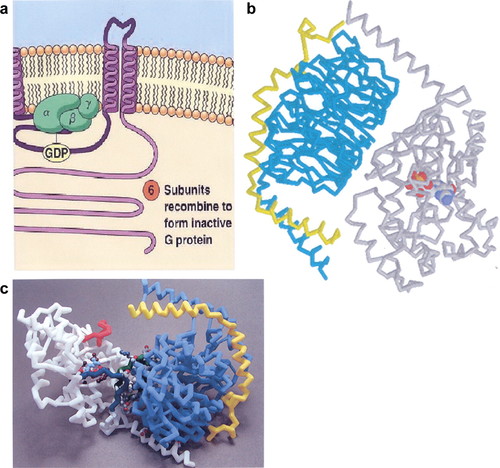
Figure 1. Various depictions of the heterotrimeric G-protein from: (a) an introductory textbook. A portion of Figure 14–5 from The World of the Cell by Becker, Kleinsmith, and Hardin © 2006, reprinted by permission of Pearson Education. The Gα, Gβ, and Gγ subunits are shown in green with a bound GDP shown in yellow; (b) an alpha carbon backbone Rasmol image derived from Protein Data Bank (PDB) file 1GOT. The Gα subunit is shown in gray, the Gβ subunit in cyan, the Gγ subunit in yellow, and a bound GDP is shown as a ball and stick representation and colored in the CPK convention; (c) as a physical hand-held model made from the coordinates in the 1GOT PDB file. The Gα subunit is shown in white, the Gβ subunit in blue, and the Gγ subunit in yellow. The bound GDP is not visible in the picture angle shown.
Advantages and Limitations of Molecular Imaging
Molecular visualization tools such as Mage, Rasmol, and Chime have been shown to increase students' understanding of molecular structure (White et al., 2002; Booth et al., 2005; see Wu et al., 2001). White et al. (2002) found that introductory biology students benefit from manipulating computer images of molecules because this allows them to get a better mental three-dimensional representation of a molecule than they would get by passively observing molecular images during a lecture. Many studies also report that students are enthusiastic about using molecular imaging software to learn about macromolecular structure (Bateman et al., 2002; Richardson and Richardson, 2002; Booth et al., 2005; Roberts et al., 2005; see also summary of previous studies in Booth et al., 2005).
When biologists use two-dimensional representations of molecules in textbooks and within computer imaging programs, they assume that all students can translate that image into a three-dimensionally accurate mental model. There are, however, documented differences in three-dimensional aptitude among individuals, perhaps most notably between men and women (Peters et al., 1995; Sorby, 1999; 2005; Sorby and Baartmans, 2000). Sorby and Baartmans (2000) found that beginning engineering male students performed significantly better on the Purdue Spatial Visualization Test: Rotation (PSVT:R) test than their female peers. PSVT:R scores were found to be significantly predicted by previous play with construction toys and drafting course work, both of which were found to be significantly different experiences between men and women. In a comprehensive review of classroom research on the link between visuospatial ability and chemistry learning, Wu and Shah (2004) conclude that low spatial ability students are at a disadvantage because they have difficulty comprehending the conceptual knowledge embedded within molecular formulas and other symbolic representations of microscopic structures.
The pedagogic value of molecular imaging programs is dependent on our students' ability to use computers efficiently and productively. Molecular imaging tools may be more useful, however, to students with superior three-dimensional skills and/or experiences. Norman (1994) states that spatial visualization aptitude is a significant predictor of interindividual variation in computer utilization performance, and that there is some evidence that students with low spatial visualization ability have more difficulty navigating computer interfaces. If we assume that students with low spatial visualization aptitude tend to be tactile learners, then students who need the most help understanding microstructure might be particularly disadvantaged when using computer imaging programs.
Multiple Representations and Learning Styles
As indicated above, students have a variety of learning styles, and the use of two-dimensional imagery of microscopic structures may be helpful to only a portion of our introductory biology students. Physical models of microscopic structures and concepts might better enable tactile learners to understand microscopic structures that cannot be touched. Many students may learn concepts better via tactile learning. Under the VARK (Visual, Aural, Read/write, and Kinesthetic) learning style framework (Fleming and Mills, 1992), these tactile learners are included in the “kinesthetic” learner category, and in the “bodily-kinesthetic” intelligence grouping in Gardner's Multiple Intelligences Theory (Tanner and Allen, 2004).
Physical Models
To overcome the inherent limitations of the computer screen or textbook pictures, rapid prototyping technology has been used to produce hand-held, three-dimensionally accurate models based on Rasmol xyz atomic coordinate data (Herman et al., 2006). Such physical models can be used in conjunction with two-dimensional images. This approach has been shown to be successful in classrooms (Wu and Shah, 2004; Roberts et al., 2005; Sorby, 2005). Wu and Shah (2004) cite several chemistry learning studies which found that students who manipulated physical molecular models had a more complete understanding of the abstract concepts underlying atomic and molecular symbols and two-dimensional imagery and did a better job of solving chemistry problems than their peers who did not use models. Sorby (2005) showed that freshman engineering students who sketched images while working with physical models in tandem with a computer imaging program as part of a spatial skills course significantly improved their score on the PSVT:R three-dimensional aptitude test and achieved higher GPA in subsequent engineering, calculus, and physics courses at Michigan Tech University than peers who had not taken the spatial skills course. One caveat to this finding is that student enrollment in the freshman course was not random, however, and so self-selection may explain some of the variation in the data. However, this study suggests that physical models can be helpful to many students.
Roberts et al. (2005) found that a class of 20 introductory biochemistry students made significant learning gains regarding their understanding of molecular structure/function after repeated use of both computer imagery and physical molecular models over several weeks. In addition, the students self-reported that the physical models were the most helpful tool facilitating their learning.
These physical molecular models, however, require a capital investment as well as a recurring time commitment from instructors to convey the unique features of each model to new student cohorts. It is important to assess the pedagogical value of tools such as these models so that instructors can make educated decisions about teaching resources.
Current Study of UW–Madison Biocore Students
To date, there have not been any controlled studies of the effect of hand-held molecular model use on students' understanding of molecular structure and function. In the current study, we measured how the use of physical models by 68 introductory biology students affected learning and attitudes as compared with a control group of 47 students who had no access to the models. These students were enrolled in the Biology Core Curriculum (Biocore) program, a four-semester Honors sequence at the University of Wisconsin–Madison (Batzli, 2005). The cell biology laboratory course (Biocore 304) is the first introduction to college-level cell and molecular biology concepts and tools for Biocore students.
In 2005, Biocore 304 instructors were given the opportunity to use three-dimensionally accurate molecular models made by 3D Molecular Designs (www.3DMolecularDesigns.com). We wanted to know whether students who used both hand-held molecular models along with computer visualization software would have an enhanced understanding of molecular structure/function relationships. We predicted that cell biology students with access to both hand-held molecular models and molecular visualization software would develop a greater understanding of how molecular structure determines function as compared with students who only had access to molecular visualization software. We also posited that access to hand-held models would increase students' enthusiasm for molecular biology, and so we also chronicled self-reported student attitudes about how these tools facilitated their learning and confidence.
To test this hypothesis, we conducted a controlled investigation of Biocore cell biology lab students in the spring 2005 semester. In all five laboratory sections, the web-based PE molecular imaging program was available to students. In the three experimental laboratory sections, the students used physical protein models for most of the semester in addition to PE. In both control and experimental groups, students' attitude and learning were assessed identically using a mixed methodology.
METHODS
Subjects and Curriculum
The subjects were sophomore undergraduate students enrolled in a cell biology lab (Biocore 304) in spring 2005. Biocore 304 is a two-credit lab course that students take concurrently with a cell biology three-credit lecture course, Biocore 303. Biocore 303/304 are the second semester courses in the four-semester Biocore program sequence. Students apply to the Biology Core Curriculum as freshmen with introductory chemistry and calculus as prerequisites and begin as sophomores. In Biocore 304 lab, an emphasis is placed on students engaging in the process of science as they learn the tools and procedures of cellular and molecular biology. During the spring 2005 semester, students designed and carried out three multi-week independent research projects in teams of four (see list of lab topics in Table 1).
| Week | Lab topic | Research action | Performance assessment | Student attitude assessment |
|---|---|---|---|---|
| 1 | Microscopy | |||
| 2 | Subcellular fractionation |
| ||
| 3 | Enzyme catalysis independent project week I |
|
| |
| 4 | Enzyme catalysis independent project week II |
|
|
|
| 5 | Enzyme catalysis independent project week III |
|
| |
| 6 | Enzyme catalysis independent project week IV |
| ||
| 7–9 | Photosynthesis independent project |
|
| |
| ||||
| 10–11 | DNA isolation, amplification & electrophoresis |
| ||
| 12–15 | Signal transduction independent project |
|
|
|
| 16 | Final exam week |
|
|
|
In spring 2005, there were five Biocore 304 lab sections, each with 21 to 24 students. Each section met for one 50-min discussion section each week followed 1 to 2 d later by a 3-h lab period. We assigned lab sections 3, 4, and 5 as the “experimental” group (n = 67; 31 females, 36 males) which had access to both physical models and the PE molecular imaging program (Martz, 2002) during the latter 12 wk of the semester. Lab sections 1 and 2 were assigned as the “control” group (n = 43; 23 females, 20 males) which had access only to PE. Our protocol was approved by the UW–Madison Education Research Institutional Review Board (protocol number SE-2004-0051). Students were given the option of not participating in the study; five students chose not to participate.
M.A.H. and Dr. Janet Batzli were cochairs of the lab course and were responsible for designing the lab exercises and for leading all five lab sections throughout the semester. Teaching assistants (TAs) conducted discussion sections, assisted the cochairs with lab instruction, and graded laboratory research reports and assignments. The concurrent cell biology lecture course met three times each week for 50 min and also had a separate 50-min required discussion section. Lecture discussion sections were independent of lab discussion enrollment, such that lecture discussions were a mixture of lab experimental and control students.
Instruction
Students had several opportunities over 12 wk to use PE and, for the experimental group, 3D Molecular Designs physical models and PE to learn about molecular structure (see Table 1). Throughout the semester, students in both groups were exposed to RasMol macromolecular images and 3D Molecular Designs physical models during Biocore 303 lectures, but only the lecture instructor manipulated the images and models during 303 class time.
Before week 4 lab, all 304 students were expected to complete the 1-Hour Tour for PE online tutorial (http://molvis.sdsc.edu/protexpl/qtour.htm) program. During week 4, lab students in all five sections were introduced to placental alkaline phosphatase structure via an individualized PE demonstration/tutorial for groups of four to eight students led by M.A.H. and R.F.P. Students in the experimental group were also allowed to see and hold alkaline phosphatase models during these in-class, instructor-led tutorials. During the remainder of week 4 lab, students were expected to work in pairs to continue their own PE investigation of alkaline phosphatase structure using a step-by-step handout written by M.A.H. and R.F.P. Students in the experimental group were also given access to alkaline phosphatase hand-held models and encouraged to use them during their PE investigation. Students in both the experimental and control groups were given bioinformatics and molecular imaging supplement handouts and were encouraged to use these for the remainder of the semester. Four sets of the alkaline phosphatase models were present in all of the experimental lab sections during weeks 4, 5, and 6, and students were encouraged to use them.
During week 7, all students were given a PE guide to photosynthetic macromolecules handout and encouraged, but not required, to investigate specific Protein Data Bank (pdb) files. During weeks 10 and 11 all students were assigned to do a self-guided investigation of transcription factor and tRNA structure using PE and to answer several questions. Students in the experimental group were also given access to transcription factor and tRNA hand-held models while working on this exercise. Similarly, during weeks 12 to 15, all students were assigned to do an individually paced PE investigation of G protein signaling macromolecule structures. Receptor and heterotrimeric G protein hand-held models were also present for students in the experimental sections.
Data Collection During Semester
Before we implemented our study protocol, graduate TAs evaluated individual performances on a standard research report summarizing the subcellular fraction guided lab exercise from week 2 (see Table 1). We considered this a baseline assessment of each student's ability to write a standard research report. During week 3, each student's three-dimensional visualization aptitude was assessed using the 20-question PSVT:R (Guay, 1977).
During semester weeks 4 to 15, a variety of learning performance assessments and self-reported student attitudes were compiled (see Table 1). Before week 4 lab, all students took a baseline survey in which students used a 5-point Likert scale to rank how well instructional tools facilitated their understanding of molecular structure/function relationships. The survey also assessed performance and their confidence in their answers (see Supplemental Material 1). The baseline survey responses were obtained at an early point in the semester when students had been briefly introduced to protein structure in 303 lectures but had not begun to study the Central Dogma and regulation of gene expression. The survey and Likert scale questions were patterned after those used by Roberts et al. (2005).
Students took a nearly identical end-of-semester survey at the end of week 15 lab. Slightly different surveys were administered to control and experimental students to reflect their respective experiences (see Supplemental Material 2a and 2b). Survey answers to the amino acid mutation question were evaluated by M.A.H. and R.F.P., who were blinded to the identities of the students as well as to whether the answer came from a baseline or end-of-semester survey (see Supplemental Material 3 for the rubric used to evaluate student answers).
During week 5 lab, teams of four to five students presented an informal PowerPoint slideshow summarizing their plans for their enzyme catalysis independent research project. Students were given feedback from instructors and peers after these presentations. M.A.H., R.F.P., Batzli, and the graduate TAs independently used a rubric (see Supplemental Material 4) to evaluate how appropriately student teams used molecular imagery and descriptions of structure/function relationships to support the biological rationale underlying their hypotheses. Individual students turned in a proposal paper, student research teams carried out their experiments, and individual students turned in a final enzyme catalysis research paper during this unit. In addition to grading these papers, TAs used a rubric to evaluate the usage of PE imagery and higher-order reasoning skills in the final enzyme paper (see Supplemental Material 5). During week 14, TAs used this same rubric to assess individual signal transduction research proposal papers.
Postsemester Interview
During week 16 (final exam week), eight control and 12 experimental students volunteered to be interviewed by S.C., who had no previous contact with the students. Interviews were videotaped. Ten females (three in control group, seven in experimental) and 10 males (five in control group, five in experimental) were interviewed. Each student was asked 17 questions about a protein that they had not studied in lab, the cytosolic portion of one nonphosphorylated receptor tyrosine kinase macromolecule (see Supplemental Material 6 for interview script). They had, however, learned about tyrosine kinase signaling pathways during lecture. At the beginning of the interview, students were shown a PE image of the nonphosphorylated tyrosine kinase as well as a hand-held model and told that they could use either or both tools to help answer any of the questions. Later in the interview, students were shown a phosphorylated PE image and model of this molecule.
Four different assessors, blinded to group affiliation, assessed the quality of answers to each question (see Supplemental Material 7a and b). M.H., R.P., and J.K. also examined interview footage to document students' tool preference and usage time while answering questions. Immediately after being interviewed each student filled out a postinterview survey (Supplemental Material 8).
Data Analysis
Students' gender, previous lecture and lab grades, three-dimensional aptitude, and baseline research paper grade were all considered as potential covariates used in a comparison of the two treatment groups (Table 2 and Table 3). Data were analyzed using the R software for statistical computing (R Development Core Team, 2008). Comparison of treatment and control groups was done using one-way ANOVA and its nonparametric alternative, the Kruskal–Wallis rank sum test. Interview questions were classified according to Bloom's Taxonomy competence levels (Bloom and Krathwohl, 1956; Yuretich, 2003) and compared with tool usage patterns (Table 4).
| Independent | Dependent-academic | Dependent-attitude | Potential covariatesa |
|---|---|---|---|
|
|
|
|
| Independent | Dependent-academic | Dependent-attitude | Potential covariates |
|---|---|---|---|
| Access to hand-held models (sections 1 & 2 = no access control, section 3, 4 & 5 = had access, experimental) | Performance on 18 oral interview questions (documented from video by 4 blinded assessors, using rubric) |
|
|
| Knowledge | Application | Analysis | Synthesis |
|---|---|---|---|
| Q5, Q18 | Q6, Q8, Q9, Q10, Q11 | Q7, Q13, Q14, Q15, Q17, Q19 | Q16a, Q16b, Q20, Q21 |
RESULTS
Assessments During Spring Semester
There were no differences between the experimental and control groups regarding their performance in the Biocore 301 lecture or 302 lab courses taken before this study, or in their baseline three-dimensional aptitude score as measured by the PSVT:R. Fifteen of the students (eight experimental, seven control) had taken a biochemistry course before the study.
There were no significant differences between the experimental and control groups for 14 of the 16 performance variables measured during and immediately after the study concluded. These variables included Biocore 303 lecture exam and final grades, Biocore 304 lab final grades, the enzyme unit final research paper, the yeast project's proposal paper, and TAs' ratings of biological rationale sophistication in the enzyme proposal and final papers. For two of the performance variables examined, the control group performed better than students in the experimental group: the mean control group student grades on the enzyme unit research proposal paper (mean = 88.6, SE = 0.97) were somewhat higher (Kruskal–Wallis P value = 0.045, t test P value = 0.062) than experimental student grades (mean = 86.9, SE = 0.49). Students in the control group also did a better job of relating their experimental data to their biological rationale in the yeast research proposal paper, the last individual assignment of the semester (Kruskal–Wallis P value = 0.033, t test P value = 0.027).
The 15 students (eight experimental, seven control) who had taken a biochemistry course previous to or concurrently during this study performed significantly better on the enzyme and yeast proposal papers (Kruskal–Wallis P values = 0.067 and 0.057, t test P values = 0.073 and 0.054, respectively), the enzyme final paper (Kruskal–Wallis P value = 0.050, t test P value = 0.061), and earned higher lab final grades (Kruskal–Wallis P value = 0.010, t test P value = 0.012) than students who had not taken biochemistry. These 15 students also wrote a more sophisticated biological rationale in their final enzyme papers (Kruskal–Wallis P value = 0.045, t test P value = 0.054) and performed better in linking their experimental results to their biological rationale in their yeast project proposal papers (Kruskal–Wallis P value = 0.015, t test P value = 0.026).
Baseline and End-of-Semester Survey Comparison
Overall, students performed significantly worse on the end- of-semester survey's transcription factor question as compared with their answer for the same question on the baseline survey (see Supplemental Material 2), regardless of whether they were in the experimental or control group (paired samples t-score = 2.1, P value = 0.04, df = 86). After responses to this question on the baseline and final surveys had been evaluated blindly by M.H. and R.P., we noticed that student answers on the final survey were generally shorter, more incomplete, or simply missing as compared with each student's effort on the baseline survey. M.H. and R.P. also agreed that the environmental conditions for the final survey were not comparable to the environment surrounding the baseline survey. Students filled out the baseline survey at the beginning of lab discussion during week 4, before addressing any other tasks. In contrast, the final survey was administered at the very end of the final lab class meeting in week 15, after students had finished presenting their final yeast research project posters and just before they left the classroom. We concluded that students' motivation, and therefore effort, on the end-of-semester survey was likely not equivalent to their effort on the baseline survey, such that any additional comparisons of these two surveys was inappropriate.
Postsemester Videotaped Interview
There was no significant difference between the 12 experimental and eight control group students who volunteered to be interviewed 1 wk after the semester ended regarding cell biology lecture final grades (Mann–Whitney U P value = 0.671, t test P value = 0.70), or cell biology lab final grades (Mann–Whitney U P value = 0.316, t test P value = 0.30). There was no significant difference in three-dimensional aptitude between the 10 females and 10 males interviewed (Kruskal–Wallis P value = 0.231, t test P value = 0.095). There was also no difference in three-dimensional aptitude scores between the 12 experimental students and the eight control students (Mann–Whitney U P value = 0.134, t test P value = 0.432).
During the interview, students either used no tool, used the model only, used PE only, or alternated between examining the model and the PE image as they answered questions. Only one of the 20 students (a control group student) appeared to use the model and PE simultaneously, and then only before this student gave an answer to Q15 and Q16 (Supplemental Material 7b).
Tool Usage and Question Type
Students chose to use the models when answering the majority of questions about a novel protein, and usage of the models increased as the question levels progressed according to Bloom's Taxonomy of competence levels (Figure 2; Table 4). Seventeen of the 20 students interviewed (85%) used no tools to answer the two “Knowledge” level questions that required recall of information learned in the concurrent 303 lecture course (see Q5 and Q18 in Supplemental Material 7b). An average of nearly 20% of the interviewees used both the models and PE when they answered questions categorized as “Application” and “Analysis,” but 54% and 60% of students, respectively, used just the models to answer the questions in these two categories. An average of 86% of the students chose to use only the models to answer the four questions in the “Synthesis” competency level, whereas none of the interviewees used PE to answer the “Synthesis” questions (Figure 2; Table 4). PE imaging usage was highest (≈13%) when students answered the five questions in the “Application” category.
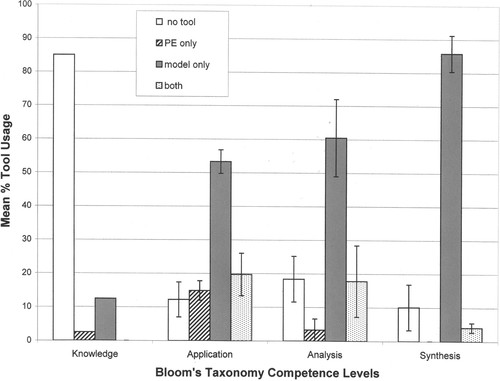
Figure 2. The average percentage of tool usage by students during the postsemester interview, regardless of experimental or control group affiliation. The 17 interview questions were categorized according to Bloom's taxonomy of competence levels (see Table 4). Tool usage during the interview was scored as either none used (no tool), PE program only, hand-held models only, or both PE and the hand-held models (both). Between 16 and 20 students' tools usage choices were recorded for each question (some students did not answer all of the questions). Error bars are ± one SE.
Comparison of Tool Usage and Answer Quality between Groups
There was some evidence that the experience with tools affected answers given to interview questions for those students in the experimental group. Overall, when any significant difference between control groups and experimental groups was identified by at least one of the four assessors, the experimental group was judged to have provided better, more sophisticated answers (Table 5).
| Question | Bloom's classification | Tool used to ID molecule? | Tool used for longer time period before answering question? | Which group produced better answers? |
|---|---|---|---|---|
| 5. Can you tell me what you've learned about protein 1°, 2°, 3°, and 4° structure? Note: 17 of the 20 students answered this question without consulting any tool. | Knowledge | Experimental (1 of 4 assessors; Kruskal-Wallis P value = 0.025, t test P value = 0.020) | ||
| 6. What kind of biomolecule do you think this is: lipid, protein, nucleic acid, or carbohydrate? | Application | Experimental group used models (Kruskal-Wallis P value = 0.088, t test P value = 0.072) | Experimental group used models (Kruskal-Wallis P value = 0.073, t test P value = 0.131). | Experimental (1 of 4 assessors; Kruskal-Wallis P value = 0.097, t test P value = 0.069) |
| 8. Can you find the N terminus? | Application | Control group used PE (Kruskal-Wallis P value = 0.091, t test P value = 0.181) | Experimental (2 of 4 assessors; (Kruskal-Wallis P values = 0.059 and 0.013, t test P values = 0.034 and 0.013, respectively). | |
| 9. Identify alpha helices/beta sheets | Application | Experimental (2 of 4 assessors; both Kruskal-Wallis P values = 0.075, both t test P values = 0.074) | ||
| 10. Does this biomolecule show any 4° structure? | Application | Experimental (2 of 4 assessors; Kruskal-Wallis P values = 0.075 and 0.057, t test P values = 0.066 and 0.071, respectively) | ||
| 16a. Propose function for colored region in biomolecule | Synthesis | Experimental (1 of 4 assessors; Kruskal-Wallis P value = 0.076, t test P value = 0.071) | ||
| 20 & 21. Propose mutations which constitutively activate/deactivate biomolecule | Synthesis | Experimental group used models (Kruskal-Wallis P value for Q20 = 0.058, t test P value = 0.046; Kruskal-Wallis P value for Q21 = 0.061, t test P value = 0.126) | Experimental (Q21) (1 of 4 assessors; (Kruskal-Wallis P value = 0.085, t test P value = 0.085) |
For 16 of the 18 interview questions, there was no difference between the experimental and control groups regarding their choice of tools (PE and/or model) when answering questions. Students in the experimental group used the model to justify their answer to Q6 (“What kind of biomolecule do you think this is: lipid, protein, nucleic acid, or carbohydrate?”) significantly more than the control students (see Q7 in Supplemental Material 7b; Kruskal–Wallis P value = 0.043, t test P value = 0.035), though none of the assessors found a difference in the quality of Q7 answers between the experimental and control groups.
Overall Comparison of Tool Usage and Answer Quality
We identified mixed results in our examination of the relationship between tool usage during the interview and answer quality, independent of whether students were in the experimental or control group (see Table 6). All four assessors did agree that 14 students who used models alone or the models + PE did a significantly better job of identifying the novel biomolecule (Q14, see Supplemental Material 7b and Table 6) than the six students who used neither tool. No students chose to use only PE when asked this question.
| Question | Bloom's classification | Tool(s) used by students who gave better answers |
|---|---|---|
| 8. Can you find the N terminus? | Application | PE only (4 students) OR models only (10 students) (1 of 4 assessors; Kruskal-Wallis P value = 0.050, F-test P value = 0.079) |
| 13. Identify amino acid sidechain residues | Analysis | PE only (4 students) (2 of 4 assessors; Kruskal-Wallis P values = 0.060 and 0.066, F-test P values = 0.187 and 0.185, respectively). Note: 2 students used only the models & 14 students used both PE and the models |
| 14. Identify this biomolecule | Analysis | Models only (12 students) OR models & PE (2 students) (all 4 assessors agreed) |
Postinterview Survey: Student Attitudes
Students who had used the models during the semester rated them quite highly as judged by an exit survey filled out immediately after the interview (Supplemental Material 8). When we asked them to “Please rate how the following tools helped to facilitate your learning of molecular structure and function this semester,” students in the experimental group rated models very highly (mean score = 4.1 out of a possible 5, with a score of 5 representing the “helped a great deal” choice). Six of the eight students in the control group appropriately chose choice 1, “never saw or touched any” for this question. The only other difference between control and experimental group responses came in their rating of the usefulness of lecture textbook readings: the control group students rated these text readings somewhat more highly (mean score = 4.3) than the experimental students (mean score = 3.7; Kruskal–Wallis P value = 0.088, t test P value = 0.194).
Both the control and experimental groups favorably rated the PE activity used during the enzyme catalysis unit, but found that using the PE program on their own was less helpful. Both groups highly rated the instructors, their peers, lecture problem sets, and prelab assignments, but gave the “physical models of biomolecules held by professors in Biocore 303 lecture” tool a less favorable rating (mean score = 3.3). Experimental group students rated the computer images of biomolecules used during lectures slightly more highly (mean score = 3.8) than control group students (mean rating = 3.3), but this difference was not statistically significant (Kruskal–Wallis P value = 0.180, t test P value = 0.192).
There was no significant difference (P value = 0.780) between the experimental and control groups in their responses to the postinterview survey question, “Which materials were most helpful to you today as you answered the interviewer's questions?” Seven students indicated that the models were the most helpful, 12 found the use of both the models and the PE program most helpful, and only one student (an experimental group member) felt that PE was most helpful during the interview.
There was also no difference (P value = 0.410) between the experimental and control groups regarding their responses to the survey question, “If you had to design a Biocore 304 research project that involved this tyrosine kinase receptor and had obtained the relevant literature, what tools would be most helpful to you?” Five students predicted that the PE program would be most helpful, two reported that hand-held models would be best, and 13 students indicated that using both PE and the models would best help them design a new lab research project.
Postinterview Survey: How Tools Helped
In the postinterview survey, students from both groups most often stated that PE was helpful to them in identifying amino acid sidechains and secondary structures, whereas the models were helpful to them in seeing the conformational changes occurring in the receptor tyrosine kinase after phosphorylation events occurred (Q2 and Q3, Supplemental Material 8).
DISCUSSION
Our hypothesis, that students with access to both hand-held molecular models and PE would develop a greater understanding of how molecular structure determines function as compared with students who only had access to PE, was supported by data we collected regarding answer quality and tool usage by 20 students who voluntarily agreed to be interviewed immediately after the semester had ended. We found good evidence that students who used the physical models in combination with the PE imaging program during the semester produced better answers to seven of our 17 interview questions (see Table 5). Our hypothesis was not supported, however, by our assessments of academic performance on typical semester assignments. There was not a statistical difference between the control and experimental groups with regards to the majority of the semester assessment pieces (both groups did equally well).
Most striking, interviewed students most often chose to use the models over no tool or PE alone in answering questions that required “higher-ordered thinking” skills (see Figure 2). Students tended not to use any tools when answering knowledge (lower-level thinking) questions. This result was the same regardless of whether students were in the experimental group or the control group. It is evident from Figure 2 that the hand-held models are very useful thinking tools and that they provide students with a resource to assist them as they contemplate complex questions, answer these questions, and as they develop new questions/ideas. This suggests then that the models are engaging and that they can be used by groups of people who have not had formal or previous training on how to use the models.
Assessments During Semester
Our data indicate that the repeated use of the hand-held models had no effect on our students' ability to do typical assignments we require in our lab course, such as pre-lab assignments, paper peer reviews, and final research papers. Contrary to our predictions, these two groups performed equivalently for 14 of the 16 performance variables we measured during the semester. It may be that our lab assignments were not sufficiently comprehensive to accurately assess the sophistication of our students' understanding of molecular structure/function relationships. Alternatively, it may be that our honors biology students were too homogeneous in terms of their potential to understand three-dimensional molecular structure and how this determines molecular interactions. This is evidenced by the relatively high scores that all of our students achieved on the PSVT:R. Bodner and Guay (1997) report that their sample of 158 sophomore students in an organic chemistry course for biology/premed majors achieved an average score of 14.2 (SD = 3.8) on the 20-question PSVT test. In contrast, our Biocore students scored an average of 15.3 (SD = 3.5) on this test. This latter explanation, however, contradicts the differences we observed between the control and experimental students during the postsemester interview.
We consider the ability to write good papers describing proposed research plans as a good indicator of scientific reasoning skills. We were thus surprised that the control group students earned higher grades on the enzyme research proposal paper, which was completed only 9 d after the alkaline phosphatase molecular models had been introduced to the experimental students. Perhaps the level of structural details displayed by the molecular models initially overwhelmed or confused students in the experimental group, such that it was more difficult for them to explain hypothetical data in terms of protein structure changes under manipulated conditions such as increased environmental temperature or pH levels. If this were the case during the enzyme catalysis research project, the experimental students seem to have overcome any such confusion a few weeks later when they completed data collection, data analysis, and wrote their final enzyme papers: we found no difference between the two groups in the final enzyme paper written 3 wk after the models were introduced.
Also contrary to our predictions, the control students did a superior job of explaining their expected data using the rationale underlying their research hypotheses in their final yeast research proposal paper of the semester. We could not compare the two groups on final individual yeast paper performance, however, because students produced a team poster at the end of the semester to summarize their completed yeast independent research projects.
Does Biochemistry Course Work Improve Cell Biology Learning?
We are confident that our experimental and control groups entered our study with comparable skills, as there were no differences in previous Biocore course grades or three-dimensional aptitude between our experimental and control groups at the beginning of this study. We did find evidence, however, that previous or concurrent biochemistry course work better prepared students to understand and apply their knowledge of molecular structure details to predict and explain experimental results. The 15 students (eight experimental, seven control) with previous or concurrent biochemistry course work experience wrote better proposal and final research papers and earned higher final lab grades than students with no biochemistry course work. These 15 students, however, were unique among typical biology students because they had begun their biochemistry course work during their sophomore year, instead of taking biochemistry as juniors after completing cell biology. Although relevant data are outside the scope of this study, their biochemistry experience suggests that they may have been advanced in other academic areas as well.
Postsemester Interview
Students who used the physical models in combination with the PE imaging program during the semester produced better answers to seven of our 17 interview questions (see Table 5). We also found that the majority of these 20 interviewees reported either the models alone or the combination of the models and PE to be the most useful when they formulated answers to questions about novel proteins, and these preferences were confirmed by the predominant usage of the hand-held models by both control and experimental students as the level of interview question increased according to Bloom's taxonomy of competency levels (see Figure 2). A comprehensive analysis of the interview data follows.
Tool Choice during Interview
Our data indicate that students tend to gravitate toward the tactile models when asked questions about molecular structure and function, even if they have had no previous exposure to the models. We documented preferential usage of the hand-held models during the individual videotaped interviews of 20 student volunteers after the semester had ended, 1 d after the students had taken their final Biocore cell biology lecture exam. Most of the interviewees preferred using the models in combination with computer imaging programs. For 16 of the 17 interview questions, control group students' tool choice was not significantly different from that of the experimental group students, and for 15 of the 17 questions, models were used the most often by all students.
We also have good evidence that students prefer to use the hand-held models to answer complex questions about macromolecular computer images and models that they have not seen before. Regardless of group affiliation, interview questions that required higher-order competencies according to Bloom's taxonomy prompted a greater usage of the hand-held molecular models (Figure 2). Conversely, most students used no tool at all when answering questions requiring only recall of facts (see Figure 2). For example, only three of the 20 students interviewed used a tool (the hand-held model) to answer interview Q18, “Dr. … spent some time in lecture telling you about receptor tyrosine kinases. Can you tell me what you know about them?” Even after we had told students that the images and model they examined during the interview were those of a receptor tyrosine kinase, students did not seem to link what they learned in lecture about tyrosine kinases with the biomolecule they scrutinized during the interview. Instead, they frequently used their hands and fingers to represent the cross-phosphorylation of tyrosine amino acid residues on two chain-like tails of aggregated receptor tyrosine kinase molecules, referring to an image used in their textbook to represent these events. This response demonstrates the influential power of the illustrations used in our textbooks and suggests that these images should be chosen very carefully to avoid student misconceptions.
Differences between Experimental and Control Groups
We did observe performance differences between our groups for seven of the postsemester interview questions, and in some cases we also saw tool preference and usage differences (see Table 5). For these seven questions ranging from “knowledge” to “synthesis” levels according to Bloom, students who had used both models and PE throughout the semester produced better answers than students who had used PE only. Questions 5, 6, 8, 9, and 10 revealed students' familiarity with molecular structure. For example, when asked to identify the type of novel biomolecule (lipid, protein, nucleic acid, or carbohydrate; Q6) they were presented with, the experimental students tended to use only the models, examined them longer, used them more often to justify their answers, and, according to one of the four independent assessors, provided better answers to this question than students in the control group. For Q8 (“Can you find the N terminus?”), experimental students provided better answers even though control group students used PE significantly longer to formulate their response. Students who had used models and PE throughout the semester also performed in a superior way on two of our highest order “synthesis” interview questions (Q16a and Q20; see Table 5). These questions required students to make predictions based on their current knowledge and the information they gathered from the physical models and PE program. The data summarized in Table 5 suggest that repeated, concurrent use of hand-held models and molecular imaging programs helps students to more accurately identify key molecular structural details on, and propose logical functions for, novel biomolecules.
Tool Use and Answer Quality
Regardless of their exposure to models during the semester, interviewed students who used models or models in combination with PE did a better job of identifying the unknown biomolecule (see results for Q14, Table 6). Students who used PE only (four students), or the models only (10 students), produced better answers to Q8 (“Can you find the N terminus?”) than the six students who used a combination of the tools. Perhaps students who used both tools were more challenged by this question and tried to use any tool available.
Results for Q13 (“Identify amino acid side chain residues”) were particularly interesting, in that the four students who used PE only to answer this question produced superior answers. This seems logical for sophomore students who are just beginning to learn biochemistry, as the PE program identifies amino acids by name when the cursor is held in place on a particular location on a biomolecule. Students using only the models would need to be familiar with specific residue structural details to correctly identify individual amino acids. One of the two students who used models only to identify sidechain residues had taken biochemistry previously.
Student Attitudes
The survey that the 20 interviewed students took immediately after their interview revealed that students' attitudes toward the use of models were very positive, and that they recognized the value of using models in tandem with the computer imaging program during the semester. The 12 experimental group students interviewed reported that the models were very helpful during the semester as they studied molecular structure and function relationships. Interviewed students also reported that use of PE during class time was helpful, but that PE was not helpful when they tried to use it outside of class. We have noticed that during lab, students prefer to ask instructors questions about the PE program rather than first exploring this program more carefully to find the answer themselves.
Although the hand-held models were seen as a good tool during lab time, students did not feel that use of the physical models during large lectures was helpful. Our lectures did use a document camera to project physical model images to the entire class as they manipulated them on stage, but apparently this is not as valued by students as having the opportunity to manipulate the models themselves.
The majority (12) of the 20 students interviewed reported that the models in combination with PE were the most valuable tools for them as they answered the questions, whereas seven of the students reported that the models alone were the most helpful tool. This preference for using both tools was also evidenced when 13 of the 20 interviewed reported that they would use both tools if they had to design a novel experiment using the receptor tyrosine kinase featured in the interview. The postinterview survey also revealed that students from both groups felt that PE was most helpful in identifying particular amino acid sidechains, whereas the models were typically cited for their use in analyzing the conformational changes that occurred in the tyrosine kinase after phosphorylation events had occurred.
Multiple Exposures to Tools
Our data indicate that students benefit from using complex tools such as the hand-held models and the computer imaging programs many times and over several weeks, and this result confirms the results of previous studies. Performance on tasks requiring visuospatial thinking has been shown to improve with frequent practice sessions requiring mental manipulation and rotation of 2D images (Lord, 1985; Tuckey et al., 1991; Sorby, 2005). Students seem to benefit the most from computer molecular imaging when they have ample time (i.e., several weeks) to learn the software and manipulate the complex images on their own or in small groups of two to three individuals (Richardson and Richardson, 2002; Booth et al., 2005).
Addressing Needs of Diverse Learning Styles
Repeatedly using multiple representations of the same microstructures may be an effective way of addressing learning style differences among our students. In their comprehensive review of the role of visuospatial thinking in chemistry learning, Wu and Shah (2004) state that, “… visual representations indeed facilitate students to understand concepts and by using multiple visual representations, students could achieve a deeper understanding of phenomena and concepts.” We feel that the models are an effective visual/tactile representation tool for our students, and our data indicate that the use of computer imaging in combination with tactile tools is a powerful instructional approach that helps our students develop the “deeper understanding” referred to by Wu and Shah. Perhaps this combination is effective because the use of both tools allows kinesthetic learners to better develop their visuospatial thinking skills.
Models as a Teaching Tool
As instructors, we frequently find ourselves picking up the models to refer to key structural details on them and on computer images while speaking about dynamic molecular interactions, particularly during our conversations with individuals and small student groups. Since we began using molecular models in our labs, our students seem to demonstrate a more sophisticated understanding of how subtle molecular structure details determine what molecules can and cannot do. These higher-order levels of learning are evidenced by the superior performance of students who had used the models with PE over the course of the semester on the interview questions that required them to apply their previous knowledge to examine the structure of a novel biomolecule. Perhaps most impressive, students accustomed to using molecular models and PE did a superior job of providing answers to “synthesis” questions that required a deep understanding of molecular structure/function relationships as compared with students who used only PE (Table 5).
We feel that the pedagogical and higher-order learning gains achieved through the use of molecular models justify the monetary investment in these tools. We would not, however, advocate using only the tactile models, as there is good evidence that molecular imaging software helps students learn about molecular structure (White et al., 2002; Booth et al., 2005; see Wu et al., 2001). Molecular imaging programs such as PE offer many instructional features that a static tactile model cannot. This was evidenced during our student interviews, where students who used only PE to identify amino acid sidechain residues on the novel biomolecule produced superior answers (see Table 6, Q13). Students' comments on the postinterview survey also showed that they recognized the value of both tools. For example, one student in the control group wrote that PE helped him in “… the identity of specific molecules that I'm not as familiar with – i.e., ligand, R-groups” while the models helped him in “… seeing the three-dimensional structure, visualizing what may happen in reactions, relations of R-groups/sidechains in the real proteins.”
Teaching as Research: Lessons Learned
Recent publications have urged instructors to study the efficacy of their instructional approaches and tools as rigorously as they would study phenomena in their labs (e.g., Handelsman, 2004; Handelsman et al., 2007), and researchers have responded with intriguing student learning data aligned with thoughtful learning objectives (e.g., Phillips et al., 2008; Robertson and Phillips, 2008). In carrying out the current study, we learned some valuable lessons about assessment tools and approaches, especially those that rely on a comparison of pre- and postperformances. For example, based on the shorter, incomplete, or missing statements that the majority of students wrote on their end-of-semester survey as compared with their more complete answers on the week 4 baseline survey, we decided that a comparison of these two surveys would not give us reliable information. We learned that the environment in which students complete assessments (quiet surroundings, time allowed for completion, etc.) and performance motivators (e.g., amount of extra credit awarded for completion) must be as similar as possible, particularly when the assessments are separated by several weeks. In other words, the most reliable student learning and attitude data are obtained when: 1) the physical environment in which students are asked to perform is as consistent as possible, and 2) every effort is made to motivate students to perform consistently and to the best of their ability.
We also learned how valuable oral student interviews can be, particularly in uncovering student misconceptions. The designers of the Biology Concepts Inventory (BCI) interview students to find out what language they use to explain complex biological processes, and use this information as well as open-ended essay questions to develop multiple-choice questions for the BCI. Our experience with student interviews leads us to heartily concur with the BCI developers' conclusion that “Listening to a student explaining ideas in a relaxed, face-to-face setting is one of the most effective means for coming to an understanding of what is “inside” students' heads, what they really mean when they select a particular distracter, how they interpret questions, etc.” (Garvin-Doxas and Klymkowsky, 2008). Educational research approaches and the resulting data are often messy but can lead to fascinating insights, especially when you take the time to sit down and have a good conversation with your students.
Future Directions
While we did not assess learning styles directly before our study, we did anecdotally note that some students were much more enthusiastic and dependent on the physical models than other students, suggesting that these students rely more on tactile learning. An intriguing future study might involve identifying learning styles among students with a tool such as the VARK, and then analyzing the addition of physical models to the curriculum. It would be important in such a study to include a student sample with greater variation in their three-dimensional aptitude, as spatial aptitude scores have been found to be a significant predictor of success for engineering students (Sorby, 1999). The implicit prediction would be that these physical models would markedly improve the understanding and attitudes of tactile learners while having much less effect with other students. A correlation analysis between learning style and attitudes might reveal relationships that are lost in studies such as ours, which emphasize group averages. In our current study, this targeted effect might also be missed given the relative homogeneity of preparation, motivation level, and abilities among our honors biology students.
CONCLUSIONS
In our controlled study we found that students who used a combination of hand-held models and molecular imaging programs over several weeks produced higher quality answers to certain higher-order questions than students who only used computer imaging programs during the same time period. These data support the findings of previous studies that reported relationships between tactile model use and student learning gains (Wu and Shah, 2004; Roberts et al., 2005; Sorby, 2005). In our study, however, the benefits to students were not found in our typical course assessments but rather were revealed during one-on-one interviews with students at the end of the semester. Remarkably, interviewed students most often chose to use the models to answer higher-ordered questions, but typically did not use any tools when answering lower-level thinking questions, regardless of experimental/control group affiliation. We feel that this is compelling evidence that the hand-held models are engaging and serve as a thinking tool for students as they determine logical answers to complex questions.
ACKNOWLEDGMENTS
Molecular models were purchased from 3D Molecular Designs using UW–Madison Instructional Laboratory Modernization Program funds. We thank Margaret Franzen and Michele Korb (Marquette University) for serving as blinded assessors of interview video-tapes. Sarah Miller (HHMI Wisconsin Program for Scientific Teaching) provided invaluable review of this manuscript. We thank Dr. Tim Herman, Center for BioMolecular Modeling, for providing assistance with understanding the molecular structural details of the hand-held models used in this study. We thank Mike Patrick for his assistance and advice in designing curricular and assessment materials. Rick Nordheim of the UW–Madison Statistics Department provided invaluable assistance with statistical analysis of our data. We are grateful to UW–Madison Biocore Program Associate Director Janet Batzli and Faculty Director Jeff Hardin for their encouragement and support of this project. Our human subjects research protocol was approved by the UW–Madison Education Research Institutional Review Board (protocol #SE-2004-0051).



