Immediate Dissemination of Student Discoveries to a Model Organism Database Enhances Classroom-Based Research Experiences
Abstract
Use of inquiry-based research modules in the classroom has soared over recent years, largely in response to national calls for teaching that provides experience with scientific processes and methodologies. To increase the visibility of in-class studies among interested researchers and to strengthen their impact on student learning, we have extended the typical model of inquiry-based labs to include a means for targeted dissemination of student-generated discoveries. This initiative required: 1) creating a set of research-based lab activities with the potential to yield results that a particular scientific community would find useful and 2) developing a means for immediate sharing of student-generated results. Working toward these goals, we designed guides for course-based research aimed to fulfill the need for functional annotation of the Tetrahymena thermophila genome, and developed an interactive Web database that links directly to the official Tetrahymena Genome Database for immediate, targeted dissemination of student discoveries. This combination of research via the course modules and the opportunity for students to immediately “publish” their novel results on a Web database actively used by outside scientists culminated in a motivational tool that enhanced students’ efforts to engage the scientific process and pursue additional research opportunities beyond the course.
INTRODUCTION
Engaging students in authentic research is an enduring recommended best practice for teaching science to undergraduates (American Association for the Advancement of Science [AAAS], 2011). Escalating national concerns about student interest and retention in science and the need for science graduates to apply concepts in creative ways have renewed the urgency for college students to move beyond being simple consumers of knowledge, to being creators of knowledge (President's Council of Advisors on Science and Technology, 2012). Like authentic research experiences, inquiry-based approaches to classroom learning emphasize the evolving process of scientific thought, provide effective opportunities for students to gain confidence in their scientific performance and see themselves as scientists, and motivate students to persist in their science education (Bauer and Bennett, 2003; Lopatto, 2004, 2007; AAAS, 2011; Harrison et al., 2011). A recent wave of pedagogical innovation has seen more instructors designing and utilizing inquiry-based laboratory exercises in their undergraduate courses as a means to engage larger numbers in the process of discovery. Ranging widely in experimental questions and techniques, these approaches have been effective at promoting self-investment; raising enthusiasm for science; stimulating student interest; and enhancing skills in experimental design, data analysis, and application of foundational knowledge (e.g., see Myers and Burgess, 2003; Howard and Miskowski, 2005; Mitchell and Graziano, 2006; Marshall, 2007; Brame et al., 2008; Siritunga et al., 2011; Treacy et al., 2011).
Although effective on many fronts, most inquiry-driven class experiences lack one critical component of a comprehensive authentic research experience: dissemination of results to the broader scientific community that will build on them. Sharing results is the important step that connects the generation of new information by a student's own hands to advancing the scientific body of knowledge—the fundamental job of a scientist. The process of disseminating results also, by necessity, sharpens required scientific communication skills. Thus, one way to significantly strengthen current research-based class experiences is for instructors to provide mechanisms to broadly share student discoveries (National Research Council, 2005) and to make the opportunity an integral part of the course.
The existing dissemination strategies are limited—most outlets for sharing data from classes are either slow, difficult to integrate into a traditional course, or do not effectively reach the appropriate scientific audience. Publication in a traditional peer-reviewed journal is one high-impact option. Articles of this type often result from class investigations that generate data of interest to a particular faculty member, and findings are included in a future publication led by that faculty member (e.g., see Malone et al., 2008). However, in this model, students are typically not involved in the entire publication process, as it often requires additional time for the faculty member to validate results and/or synthesize student contributions, thus delaying the submission until long after the end of the course.
To reduce this degree of dependence on a faculty member, many journals specific for publishing undergraduate work have come online over the past 15 yr (Tatlovic, 2008). Some of these journals are peer-reviewed and broadly distributed, including the Journal of Young Investigators (www.jyi.org), BIOS (www.tri-beta.org/bios.html), and the American Journal of Undergraduate Research (www.ajur.uni.edu). A few offer powerful additional learning opportunities, like the Journal of Young Investigators, which includes students in every step of the peer-review process. While the great strength of this type of publication mechanism is that responsibility is put on the student to see the process through, this can also be a limitation. As the turnaround time from submission to publication is typically no less than 2 mo, these options often require a time commitment well beyond the end of the course. With student follow-through challenged by competing interests and commitments, most end up missing this potentially transformative final step in the research process. One strategy to deal with these limitations is to build journal publication into the structure of the course (Jones et al., 2011), but this works only if review of student submissions begins within a few weeks after the start of the class—a very short time frame that leaves little time for experimental repetition and validation of results before submission.
The ease with which material can be published directly on the Internet has created several new opportunities for dissemination of undergraduate research. Such formats are more immediate than traditional means of publication, owing to greatly reduced or no outside peer-review component. One common form is an open clearinghouse covering a variety of fields, typically in the form of institution-based websites for local on-campus research. Others, such as the National Undergraduate Research Clearinghouse (www.webclearinghouse.net), accept faculty-sponsored data from students across the country. Although sites of this nature make dissemination of students’ full reports feasible within the time frame of a course and can be easily utilized by course instructors, they generally have low visibility among professional researchers and rarely reach the scientists who are most likely to use and build on the findings.
Web-based strategies are also used by larger consortiums that involve classes of students in research that is united by common goals. Examples are initiatives such as the Genomics Consortium for Active Teaching (Campbell et al., 2006, 2007), Genomics Education Partnership (GEP; Lopatto et al., 2008; Shaffer et al., 2010), and SEA-PHAGE (Howard Hughes Medical Institute, 2011). Some of these very successful programs have built-in means for dissemination of research outcomes (e.g., GEP: http://gep.wustl.edu/projects/data_repository). While these efforts are popular and provide well-rounded, authentic research experiences for students, most leave little flexibility for how students report their discoveries. Results are often synthesized into specific output formats, a convention that limits student practice with presentation of their data, data interpretation, and their novel scientific conclusions.
We present a modified dissemination model developed with the goals of: 1) broadening the dissemination experience to more students doing a range of class-based research and 2) increasing the impact of student generated findings by publishing them directly to an appropriate scientific user group. The identification of an appropriate user group and development of an efficient means of sharing results with them are described. In short, students were given the opportunity to perform novel genome structure and function studies in a sparsely annotated genome and to present their findings in a Web database. We then leveraged these new data to connect students and their results with the “user group” of scientists in need of their results, by updating the community's official genome database and linking to the students’ reports. The Web format allowed students to make immediate, tangible, and credited contributions to a vital community resource within the duration of a single college course. Changes in student behaviors and attitudes toward engaging research positively correlated with the opportunity for immediate Web publication directed to an interested audience in two molecular biology courses that were assessed over 5 yr.
METHODS
Developing Research Opportunities for Courses That Fulfill Community Needs
We reasoned that coupling classroom research experiences with dissemination directly to a “user audience” would enhance students’ investment in the science, sense of empowerment, and motivation to effectively communicate their results. Opportunities for participation in annotation of genome structure and function in a newly sequenced organism, the ciliate Tetrahymena thermophila, provided the basis for this type of experience. Tetrahymena is one of two major ciliate model systems, along with Paramecium. It is amenable to a wide array of laboratory techniques that allow researchers to study a variety of basic cell biology processes (Collins and Gorovsky, 2005). Since the sequencing of this species’ genome in 2003, the community of ∼300 researchers worldwide has been served by a model organism database (MOD) called the Tetrahymena Genome Database (TGD: http://ciliate.org;Stover et al., 2012). In 2007, TGD began development of a new wiki interface, making it one of a small number of MODs that allows members of the research community to directly annotate the genes on which they work. This format allows users to upload annotations based on unpublished data, which are desired by the TGD user community, as about 70% (∼17,000) of the genes in this evolutionarily isolated organism have no functional assignment (they are labeled “hypothetical protein” in the current annotation). The Tetrahymena community values these efforts to provide much-needed information on gene structure and function. We therefore took the opportunity afforded by the structure of TGD to facilitate dissemination of discoveries by our undergraduates.
Designing Inquiry-based Research Protocols
Modules guiding investigations of Tetrahymena genes were developed explicitly for integration into existing laboratory components of standard college courses in molecular biology, introductory biology, and cell biology. The modules guided discoveries toward understanding gene function in several areas of study: gene expression throughout the life cycle or in response to different stresses, localization of encoded proteins, in silico analyses of DNA and protein sequences, and analysis of gene knockout phenotypes. Importantly, students also tested computationally predicted gene models (introns, exons, and start/stop codons), which are often miscalled in genome-sequencing projects and are in need of empirical editing. Because the Tetrahymena genome is relatively large and unannotated, students at different stages and in different fields are able to make significant contributions.
Developing a Database for Dissemination of Student-generated Experimental Results
A Web database prototype was created by two undergraduate students at the Claremont Colleges. It was designed to allow uploading of student data and accompanying text by individual instructors from any institution participating in class-based genome annotation. Students engaging investigations for functional annotation of Tetrahymena genes (through the class research modules discussed above) reported their findings via the database. Users could sort the uploaded data and reports by gene accession number, gene name, and a few common phenotypes. The database resource was advertised to the Tetrahymena research community through publication (Smith et al., 2012) and platform sessions at the semiannual Federation of American Societies for Experimental Biology Ciliate Molecular Biology conference, the premier conference for ciliate biologists. During this meeting, genes of broad interest were solicited for students to study and were prioritized with input from the larger research community. The database was designed such that multiple students could report new information for the same gene, thus modeling for participants how scientific knowledge is constructed over time, a common learning objective that is challenging to meet (Hunter et al., 2007).
The most recent version of the database for student results, called SUPRDB (Student/Unpublished Results Database: http://suprdb.org), was developed by a team of programmers at Bradley University and focuses on usability and flexibility for integration with MODs. SUPRDB allows users to upload reports written in standard scientific format (abstract, introduction, methods, etc.) and accompanied by data images generated over the course of scientific experimentation. Interfaces also exist for providing links out to community stock centers and Gene Ontology annotations. Each report receives a unique SUPRDB identification number (SUPRDB ID) that can be used to link to the study from other websites. The TGD Wiki was modified to accept identifiers and show links to SUPRDB reports alongside links to papers indexed by PubMed. Users are encouraged to update the annotations shown for genes at TGD Wiki based on results in SUPRDB, making these reports a valuable and important source of support for the information displayed at TGD Wiki. Importantly, these links from TGD Wiki have maximized accessibility and visibility of student discoveries to the broader ciliate research community.
Study Design
Two groups of students (referred to as “experimental” and “control”) were compared. Students in both groups were from two molecular biology courses taught multiple years by the same instructor in the Keck Science Department of the Claremont Colleges (Table 1). The course-based research experience for each group was nearly identical in structure: both groups investigated protein localization and gene expression, guided by the same research modules, with each student pair investigating a different protein/gene. In addition, the class size, the laboratory facility, and the instructor were constant between control and experimental groups. The comparative study was conducted over 5 yr of students engaging the research modules in these courses. Students in the first 2 yr of the study were the control group—no online database had yet been created for the project; they were not offered the opportunity for immediate Web publication of their discoveries (n = 44; Table 1). This group was told that their results would likely be made available to the larger research community at some time in the future. Students in years 3–5 were the experimental group—they were offered the chance to immediately publish their discoveries via the database, which would be accessed by the broader ciliate research community (n = 95; Table 1). Students in both groups were a mix from three small, highly selective liberal arts colleges: Claremont McKenna (C), Scripps (women's college, S), and Pitzer College (P). Analyzing the percentage of students from each college participating in the courses using a Student's t test for differences between the means revealed no significant differences in college affiliations of students in the two groups (Table 1). In both groups, the percentage of students from Pitzer College were the minority, while the percentage of students from Claremont McKenna and Scripps Colleges was almost equivalent. There was a difference in the percentage of male and female students between control and experimental groups (p < 0.05 with Student's t test). However, in each group, there were significantly more female than male students (Table 1). Considering these parameters, the only major structural difference was the experimental group being offered the additional opportunity to Web-publish their discoveries to a target audience.
| Academic year | Course | Number of students | % F, Ma | % C, S, Pb |
|---|---|---|---|---|
| Control group (no database publication) | ||||
| 2005–2006 | Bio 170Lc | 18 | 67, 33 | 44, 44, 12 |
| 2006–2007 | Bio 170Lc | 18 | 55, 44 | 50, 33, 17 |
| Bio 173Ld | 8 | 63, 37 | 38, 50, 13 | |
| Total | 44 | AV: 62, 38 | AV: 44, 42, 14 | |
| Experimental group (+ database publication) | ||||
| 2007–2008 | Bio 170Lc | 18 | 72, 28 | 44, 50, 6 |
| Bio 173Ld | 10 | 70, 30 | 50, 40, 10 | |
| 2009–2010 | Bio 170Lc | 19 | 79, 21 | 32, 50, 18 |
| Bio 170Lc | 17 | 82, 18 | 35, 47, 18 | |
| 2010–2011 | Bio 170Lc | 18 | 78, 22 | 34, 44, 22 |
| Bio 173Ld | 13 | 85, 15 | 46, 39, 15 | |
| Total | 95 | AV: 78, 22 | AV: 40, 45, 15 | |
Students were introduced to their research project and goals on the first day of class. In both experimental and control groups, this included an introduction to the Ciliate Genomics Consortium of faculty and students, to which they would automatically belong by engaging in the research. They were also introduced to the TGD, the source of gene information for their studies. Emphasis was placed on how their particular research would fit with the broader research initiatives of the community. Both groups were offered opportunities to work outside class time through arrangement with the instructor. The fact that working outside class was completely optional and would have no bearing on their final grade was made clear—students individually acknowledged their understanding. The experimental group was additionally introduced to SUPRDB (formerly Ciliate Genomics Consortium Database), which links directly to and from gene pages at the TGD Wiki to make student results immediately accessible by the research community. The key role that Web publication on SUPRDB would play in sharing their results with the user community was emphasized. Students in both groups understood that, although they might not have enough time to produce journal publication–quality data, the chance that future students would continue the work or repeat and validate their results was high.
Assessments and Data Collection
The in-class research projects spanned 10 wk, filling one 4-h laboratory session each week. Data for each class were collected for 1 yr, beginning at the start of the research project.
Motivation Assessment.
Motivation to engage research was assessed by collecting data on research engagement behavior (primarily time on task). Students were given the option of extending their work on the research activities beyond the allotted class time. The number of students who voluntarily took this option and the duration they engaged the research outside class time was analyzed.
Data Collection. On any single day, the instructor supervising the work recorded the names of those students opting to continue their investigatory work on the project outside class time and the amount of time each student worked in the laboratory to the nearest 15-min interval. “Work on the project” included making reagents, designing and executing experiments, making/recording observations, and updating notebooks. Time spent on report writing was excluded from the data set, as were other “homework” types of activities. The supervision and data collection rotated between one of two course instructors and a teaching assistant. In addition, the names of students electing to work on any part of their project beyond the conclusion of the entire course, into the next semester or summer, was tracked by recording requests for assistance from one of the instructors, which was necessary for continuing the work. In this case, those students working on report writing were also included. Motivation to pursue additional research beyond the course was also assessed. The names of students in each class who pursued an additional research opportunity within 1 yr following the conclusion of the course were obtained through a survey sent to all students 1 yr out from their in-class research experience. The question yielding this data was: “Have you pursued another research experience (not counting thesis) after the end of the course titled ‘Molecular Seminar’ or ‘Molecular Biology’ that you took last year? Y/N.” These data were also obtained by students directly contacting one of the instructors to request letters of support for research position applications.
Data Analysis. For each measure, an average of the data from each course in the control and experimental groups was calculated as a percentage of total students. The course average percentages were then used to calculate an overall mean percentage for the control group and the experimental group, and were further analyzed by a two-tailed Student's t test for statistical differences between the overall mean percentages.
Scientific Communication Assessment.
Outcomes related to scientific communication were assessed with a rubric for grading the scientific format research reports submitted by students for a grade at the conclusion of the project, before the end of the course (see the Supplemental Material). Although students had the option of continuing their work on the report after the conclusion of the course, the reports were not regraded at that point; thus, all data were from original submissions of their reports within the time frame of the course. The reports submitted in control and experimental groups were scored by the same instructor, utilizing the same rubric (with only slight modifications over time). Scores for each rubric section of each student's report were recorded on a master spreadsheet for that course.
The individual scores for each report section were averaged for each class in the control and experimental groups. Averages were then used to compute an overall mean for each group, and then further analyzed by a two-tailed Student's t test for statistical differences between the means.
Anecdotal Attitudinal Assessment.
Attitudes about the publishing opportunity were gathered from standardized anonymous student evaluations designed by the institution and administered at the conclusion of the course in the absence of the instructor (but with student assistance). An open-ended, free-response section that allowed for targeted questions from the instructor was used to request student feedback on the in-class research experience and the opportunity to publish in the database. Students were asked to comment specifically the authentic research experience with opportunity to publish that they experienced—any impact (positive or negative) on their attitudes toward doing science or motivation to engage course lab work was solicited. They were also asked to suggest ways to improve the experience. Resulting comments were analyzed by the instructor(s), one being constant between all courses. Comments were scored according to a five-category rubric for the impact of the publication opportunity as follows:
Category 1: Positive impact. Comments reflected that publication opportunity increased the student's motivation to engage in the course research or future research, or improved the student's attitude toward doing science. | |||||
Category 2: Neutral. Comments reflected that the opportunity to publish had no effect on the student's motivation to engage or attitude toward doing science. | |||||
Category 3: Negative impact. Comments reflected that the opportunity to publish diminished the student's motivation to engage the course-based research or created negative feelings toward doing science. | |||||
Category 4: Comments specific to the impact of the publishing opportunity dimension of the research experience cannot be separated from positive comments about the entire research experience as a whole. | |||||
Category 5: Comments specific to the impact of the publishing opportunity dimension of the research experience cannot be separated from negative comments about the entire research experience as a whole. | |||||
RESULTS
Student Research Results Improved Genome Annotations
Results from student investigations were immediately disseminated to the research community in two ways: 1) uploading data and reports to an unpublished results database (currently SUPRDB) and 2) contribution of annotations to TGD Wiki based on the data students collected. There were 130 student-authored reports uploaded to the results databases over 3 yr. Approximately 25 reports addressed protein localization, 40 addressed gene expression, five reported knockout phenotypes, 12 identified errors in gene models by comparing empirically derived base sequences of both genomic and cDNA copies of the gene, and 80 reported discoveries derived through in silico methods. In many cases, these reports described multiple experiments for each gene and reported findings on different aspects of protein or gene characterization. Students used their results to designate an official name for the gene they investigated, one that reflected its putative function based on their research. These names were listed in both the student results database and in TGD Wiki, allowing mutual links to be created between the two databases. TGD Wiki was further updated by editing the short functional description given for each gene. Thirty genes also received contributions to the general information section of the gene pages that provided in silico–based analyses of the gene models or extended functional descriptions (for examples, see the genes MAF4 and STF1 at www.ciliate.org).
Dissemination Opportunity Increased Student Motivation to Engage Science
The outcome from disseminating student results to a target audience on student motivation to engage the research process was assessed by science-engagement behaviors in two courses at the Claremont Colleges: Molecular Biology (Bio170L) for juniors and seniors and Molecular Seminar (Bio173L) for sophomores (see Methods and Table 1). Findings from comparisons between students engaging the research modules in cohorts before, versus after, database publication was available are summarized in Table 2. The opportunity for database publication correlated with: 1) a 49% increase in the number of students electing to work outside class and a fourfold increase in the average number of voluntary hours students spent outside class working on the project; 2) a 57% increase in number of students voluntarily working on aspects of their projects (including refinement of their project reports) beyond the duration of the course; 3) a 12% increase in students who pursued research opportunities in the following year; and 4) a 21% increase in average quality scores of laboratory reports, judged according to a grading rubric (see the Supplemental Material), with the most improvement observed for data presentation and analyses in the results section and depth of discussion content (Figure 1).
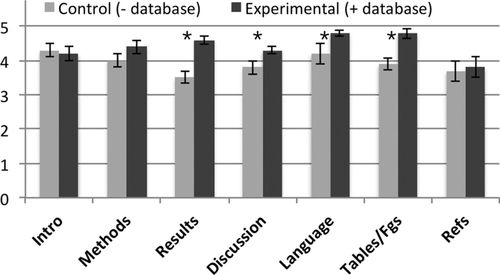
Figure 1. Scientific writing outcomes. For control and experimental groups, average course scores were compared for each category on a grading rubric (see the Supplemental Material). “Intro,” introduction; “Methods,” materials and methods; “Tables/Fgs,” tables and figures; “Refs,” references. For control group, n = 3 courses; for experimental group, n = 6 courses. Asterisks indicate statistically significant differences (p < 0.01) when compared using a Student's t test.
| Motivation measure | Control (no database publication) | Experimental (+ database publication) | Difference |
|---|---|---|---|
| Percent students working outside class time (± SD) | 35 (± 6) | 83 (± 9) | + 48%** |
| Number of hours outside class, per student, per week (± SD) | 0.5 (± 0.1) | 2.2 (± 0.46) | + 440%** |
| Percent students working beyond end of course (± SD) | 2 (± 1) | 61 (± 15) | + 59%** |
| Percent students pursuing more research within 1 yr (± SD) | 54 (± 7) | 67 (± 6) | + 13%* |
Anonymous student feedback collected from institution end-of-term evaluations was also encouraging. Evaluations reflected positive attitudinal outcomes from the opportunity to Web-publish discoveries to interested scientists (n = 79 student comment forms). Using a comment-scoring rubric (detailed in Methods), we found 54% of the student comments reflected “positive impact” (category 1); 8% were “neutral” (no detectable impact; category 2); 0% reflected “negative impact” (category 3); 36% were positive comments about the research experience as a whole, wherein the contribution of the publishing opportunity alone was undistinguishable (category 4); and 2% were negative comments about the research experience as a whole, but the contribution of the publishing opportunity alone was undistinguishable (category 5). The following few quotes represent positive impact and a range of outcomes for students at different levels. From a second-year student: “The chance to web publish emphasizes the confidence [Dr. X] has in our work. This appreciation and held esteem allows us to thrive as future scientists and inspires us to seek answers to our own areas of uncertainty.” Another second-year student commented: “Publishing our results made me feel like a real scientist. In fact, I realized that I had become one.” A senior notes: “Knowing that other scientists will see our results and possibly use them for their own research is extremely exciting (and scary). I look forward to doing more research in the future!” A comment scored as neutral was from a senior: “The lab was interesting, but I didn't really care if I published or not since publication will probably not be a part of my future in medicine.” Overall, no adverse effects of immediate data publication were evident from the comments.
DISCUSSION
Our data revealed a positive correlation between Web-publication opportunities and student motivation to engage the scientific process. Students voluntarily spent more time on task, learning and practicing science, which included designing, performing, and repeating experiments, as part of class-based research experiences when a mechanism to publish their original findings to an interested scientific community was an integral part of the course. Students also spent more time analyzing, organizing, and presenting their data, and developing scientific writing skills to communicate their results in scientific format. A greater number pursued additional research opportunities. We recognize that there may be other variables that could account for differences between the comparison groups that are difficult to rule out, such as the instructor improving over time with the course or more options for research outside class being available during some years. However, anonymous student comments from standard course evaluations reflected positive impact from the targeted database publication opportunity. As a side note to this study, a separate set of assessments revealed that 56% of first-year students who published their findings on the TGD Wiki (from work done in their introductory biology course) pursued additional research opportunities within 3 mo following their publication to the database. This fraction was markedly higher than seen with cohorts who did not have the research/publication experience, but we lack a control group that would elucidate the contribution of only the publication opportunity.
The model for class-based research with immediate publication described here is broadly transferable to other teaching settings in the following ways:
The gene annotation research aspect of the model can be applied to other model-system communities with genome annotation needs to simultaneously benefit both students and the research community. For class research curricula developed to explore gene structure or protein function, our model illustrates how a database for student results may be created and linked to an official genome database for an organism. This effort required coordination with the genome database staff and education of the model organism community about the source of this student-generated data, its strengths, and possible limitations. TGD Wiki is one of the few MODs that, rather than employing a curation staff, relies entirely on the research community to maintain functional annotations at the website. Other notable MODs that follow this model are those for Escherichia coli (EcoliWiki: ecoliwiki.net/colipedia/index.php) and Bacillus subtilis (SubtiWiki: http://subtiwiki.uni-goettingen.de). However, many other MODs now have some type of community annotation portal for researchers to submit annotations to the curators or upload supplementary annotations, including SGD (http://wiki.yeastgenome.org), WormBase (http://wiki.wormbase.org), TAIR (www.arabidopsis.org/doc/submit/functional_annotation/123), and MGI (www.informatics.jax.org/submit.shtml). The importance of, and reliance on, community annotation is likely to increase as the production of genome sequences accelerates beyond the funding to provide annotations (Howe et al., 2008; Mazumder et al., 2010). This trend is being seen even at established MODs, such as TAIR, whose declining funding situation (arabidopsis.org/doc/about/tair_funding) and potential for community involvement have been well documented (Berardini et al., 2012). We thus see many possibilities for other model system communities to embrace genome annotation by undergraduates, and these should only grow as resources become more limited. Developing a cohort of young scientists with experience performing functional annotations will also help with this challenge in the future. These considerations imply that genome-based student research models with means for targeted dissemination have strong future potential to benefit a wide range of model organism research in ways that mutually benefit undergraduate education.
The modules developed to guide studies on Tetrahymena gene function may be used with students in a variety of courses at any institution, and student findings from these studies may be published on SUPRDB. To date, more than 500 students from nine institutions (state universities, private universities, and liberal arts colleges) have used the Tetrahymena genomics research modules to investigate functions of ~300 Tetrahymena genes. More information on the research consortium and how to get involved is available on the Ciliate Genomics Consortium website: http://tet.jsd.claremont.edu.
The SUPRDB database for student/unpublished results is available for anyone to use as a platform to publish student data that support functional gene annotation, and it allows posting of reports in a multitude of languages. The interface includes features designed for maximal integration and utility with existing MODs, providing a stable URL to each report, a clearly marked section showing Gene Ontology annotations supported by the work, and links to the community stock center for accessing strains and constructs described. Though the database is currently tailored for integration with TGD, the schema and programming may be obtained from developer Nicholas Stover (Bradley University) for hosting on another server, then modified to meet the specific needs of other genomic research communities. Alternatively, the current database and website hosted by Bradley University may be modified to accept data and share links with genomic databases from any interested community.
The immediate publication model is not without its challenges and potential pitfalls. Creating the opportunity for Web publication requires that an instructor be ready to provide the time and support necessary to acquire data that both student and instructor are satisfied with publishing in the database. Using the example of the Tetrahymena research community, it is generally accepted that these student-generated data will not be flawless, but attempts to validate results and apply rigor are expected. Accordingly, students often request time outside class to work or into the following academic term (or summer break) to complete aspects of their study to a point at which they are comfortable publishing, which is decided in close consultation with the instructor. In total, this may require more instructor time than originally budgeted for course activities; the challenges of guiding student efforts in experiment execution and writing for publication can be significant. It may be worth noting that in recent iterations of the course-based research in the two molecular courses described, the research report grading rubric (see the Supplemental Material) served as criteria for report publication in the database—only those reports scoring 4 or above in each category and a 4.2 average overall were posted in the database. Approximately 15% of the reports did not meet this minimum criteria in the first graded submission, and 10% were never improved or published.
Instructors have addressed the problem of limited time for data validation in different ways. One approach is to limit experimental time to only the class period, then ask students to evaluate their confidence in the results they have at the end of the term, using a rating scale (e.g., 1–10) or to rate their data as “soft,” “somewhat firm,” or “solid.” Questions that guide students in these evaluative exercises prompt them to think about possible sources of error, what could be done to minimize error, alternative ways the data could be interpreted, and the importance of ruling out these other interpretations. The ratings and their justifications may accompany the data posted by the student. Typically, the ratings also include suggestions for further validation of the results. For example, students have rated an experimental result as soft if it was done only once or did not have the best controls to rule out alternative interpretations. Peer review has also been used for data evaluation, with one student's experiment and data being evaluated by at least one other student and the students working together to propose a rating and justification. Critical evaluation by both self and peers in this context has been leveraged as a powerful learning opportunity that simultaneously provides important information about the Web-published data to the larger science community. Although this strategy could be used in any class in which students generate original data, the opportunity to publish requires that evaluation be taken seriously and highlights the importance of critical evaluation in the publication process. Web-publishing ratings and rationales along with data allows sharing of results in various states of validation by the end of the course, as outside researchers then have a more complete picture of experimental rigor and data validity.
The Tetrahymena research community has been strongly supportive of this education initiative and its integration as a community resource. A couple of major concerns have been addressed in the process. Questions about validity of student-generated results were addressed in the ways described, and the community seems satisfied with the current degree of instructor oversight and internal data evaluation. Another concern was about copyright issues—some journals do not allow publication of data posted on other websites. We agreed that, at any time, a student author or supervising faculty member could request the curator remove their data from the database/website. This strategy has been used successfully by another database to address the same issues (National Undergraduate Research Clearinghouse: www.webclearinghouse.net).
In sum, our engagement-behavior data and scientific writing data suggest that immediate dissemination of student results to a popular community resource can heighten the efficacy of course-based research by motivating student engagement with all aspects of the scientific process, including critical evaluation of their own data and that of others. With MODs emerging as important new outlets for members of the scientific community to share their findings, opportunities to increase impact of undergraduate research have also arisen. Enlisting students to contribute to these resources is mutually beneficial and ensures that the data they collect can be used by subsequent students and other researchers for years to come.
ACKNOWLEDGMENTS
We gratefully acknowledge Douglas Chalker and Joshua Smith for their critical contributions to building the Ciliate Genomics Consortium and thank J. Smith for assisting with data collection. We also acknowledge the support provided by the Bradley University Center for STEM Education. This work was supported by a CAREER award from the National Science Foundation to E.A.W. (MCB 0545560).



