Beyond Punnett Squares: Student Word Association and Explanations of Phenotypic Variation through an Integrative Quantitative Genetics Unit Investigating Anthocyanin Inheritance and Expression in Brassica rapa Fast Plants
Abstract
Genetics instruction in introductory biology is often confined to Mendelian genetics and avoids the complexities of variation in quantitative traits. Given the driving question “What determines variation in phenotype (Pv)? (Pv=Genotypic variation Gv + environmental variation Ev),” we developed a 4-wk unit for an inquiry-based laboratory course focused on the inheritance and expression of a quantitative trait in varying environments. We utilized Brassica rapa Fast Plants as a model organism to study variation in the phenotype anthocyanin pigment intensity. As an initial curriculum assessment, we used free word association to examine students’ cognitive structures before and after the unit and explanations in students’ final research posters with particular focus on variation (Pv = Gv + Ev). Comparison of pre- and postunit word frequency revealed a shift in words and a pattern of co-occurring concepts indicative of change in cognitive structure, with particular focus on “variation” as a proposed threshold concept and primary goal for students’ explanations. Given review of 53 posters, we found ∼50% of students capable of intermediate to high-level explanations combining both Gv and Ev influence on expression of anthocyanin intensity (Pv). While far from “plug and play,” this conceptually rich, inquiry-based unit holds promise for effective integration of quantitative and Mendelian genetics.
INTRODUCTION
Teaching and learning of genetics in many introductory biology courses is typically limited to meiosis and analysis of Mendelian inheritance of discrete traits using Punnett squares. While Mendel's work has great historic significance, and working problems focused on discrete traits has classic relevance, the traditional genetics curriculum needs updating and should mirror the work and problems undertaken by 21st-century geneticists. Specifically, both Dougherty (2009) and more recently Redfield (2012) make the case for integrating or even “inverting” the genetics curriculum, so that the historical approach of teaching meiosis and inheritance of single-gene Mendelian traits first and foremost is reversed and the curriculum leads with inheritance and expression of common, recognizable, and measurable continuous traits (also known as multigenic or quantitative traits, e.g., height, color, size) influenced by multiple genes and environment. In other words, a new curriculum could lead with questions and inquiry into quantitative genetics and integrate Mendelian genetics along the way. Moreover, folding in concepts, language, and mechanisms of evolution within an integrative genetics unit like the one reported here makes for a fluid curricular transition from genetic inheritance to the complexities inherent in the teaching and learning of evolution.
To meet the challenge of curriculum development, we used plants as our model system, more specifically, rapid-cycling Brassica rapa (RCBr) (Williams and Hill, 1986), also known as Fast Plants, to allow for flexible and low-cost inquiry into quantitative genetics. We developed a unique 4-wk guided-inquiry laboratory unit in which students investigate the inheritance and expression of anthocyanin pigment intensity of RCBr grown in various environments. The Wisconsin Fast Plants program (www.fastplants.org) has developed many genetically distinct stocks of RCBr, including those that express anthocyanin pigment intensity as a quantitative trait (shades of purple from high to low) that is heavily influenced by environmental conditions. Additionally, stocks have been bred to carry the anthocyaninless (anl−) allele that exhibits simple Mendelian inheritance and results in green plants that lack anthocyanin (homozygous recessive) and purple plants (heterozygous or homozygous dominant). In short, anthocyanin inheritance and expression in RCBr can be studied from the perspective of both quantitative and Mendelian genetics.
Why use Fast Plants for studying genetics and evolution in an educational setting? Fast Plants predictably grow from seed to seed in 40 d under a range of classroom environments. Importantly, from a genetic and evolutionary perspective, Fast Plants are self-incompatible, thus insuring allelic variation that conditions a wide range of phenotypic variation within populations. Hundreds of Fast Plants can be grown in a small area, and seed can be stored for up to 20 yr, making it possible to grow and compare multiple populations and generations with unique genetic heritage at the same time. Furthermore, Fast Plants’ growth, development, and reproduction exhibit a high degree of phenotypic plasticity in response to varying environments, and the traits are quantifiable. The Wisconsin Fast Plants Program has developed many rapid cycling, genetically distinct stocks of six interrelated Brassica species, including rapid-cycling RCBr, that are catalogued in the Rapid-Cycling Brassica Collection (rcbc.wisc.edu). With modest investment in simple lighting setups and carefully selected seed stocks, students can develop and test sophisticated questions within an inquiry-based genetics unit such as the one presented here.
The unit we designed integrates quantitative and Mendelian genetics together with concepts of variation, selection, and genetic drift into a large-enrollment introductory-level Ecology, Genetics, and Evolution laboratory course. The primary learning outcomes for the unit were for students to 1) develop and carry out an experiment to provide evidence about how variation in phenotype is influenced by variation in genotype and the environment and 2) use data as evidence to explain inheritance and expression of a continuous (quantitative) trait.
Genetics is challenging to learn and teach, given the complexities of language (Cho et al., 1985; Helm, 1991), transitions between macro-, micro-, and molecular-level scales (Marbach-Ad and Stavy, 2000; Dauer et al., 2013), and persistent misconceptions (Bishop and Anderson, 1990; Anderson et al., 2002; Smith et al., 2008; Andrews et al., 2012). Study of genetics brings forth a particularly large and complex set of concepts, including deep and broad understanding of entities and processes that create, disrupt, transmit, and maintain variation of traits from micro- to macrolevels, across generations, and from cells, individuals, and populations (Stewart and Van Kirk, 1990; Duncan and Tseng, 2010). As undergraduate students begin their study, most have been exposed to genetics and evolution concepts and terms through prior course work, everyday experience, and popular media. Even at the basic level, students form a mental framework or cognitive structure for how their ideas, and representations of ideas in language, relate to one another (Deese, 1965); however, their conceptions are usually not completely developed, accurate, or precise (Marbach-Ad and Stavy, 2000; Marbach-Ad, 2001).
Conceptual change theory of learning proposes that students bring their prior knowledge and familiarity with concepts and language to the classroom, laying the foundation for wrestling with new concepts; replacing, reorganizing, and revising understanding, and constructing new knowledge (Posner et al., 1982). We used conceptual change theory, combined with the notion of threshold concept learning as a heuristic, to begin to examine the development of students’ mental frameworks or cognitive structures and their understanding during this new integrated genetics unit. Threshold concepts and transformational learning stems from conceptual change learning theory and provides a potential framework for examining learning in the classroom (Meyer and Land, 2003, 2006). According to Meyer and Land (2003) and a growing literature base,1 threshold concepts are domain specific, yet broad, and frequently difficult for students to grasp, but when understood, they work as gateways or portals for deeper, holistic understanding of a whole web of concepts within the discipline. Taylor (2006) and Ross et al. (2010) proposed some specific threshold concepts in genetics and evolution that include variation, randomness, scale (temporal and spatial), and uncertainty. We focused on variation as a potential threshold concept that can be examined at all scales of biological organization, through mechanisms that promote or maintain variation and change variation over evolutionary time. Given that each student constructs genetics knowledge and understanding from a foundation of prior knowledge that has structure and associative meaning for the learner (Shavelson, 1972), we sought to examine students’ cognitive structures combined with the notion of threshold concepts and to build an instructional unit that integrated basic concepts with the complexities of variation, specifically phenotypic variation (Pv) as conditioned by variation in genetics (Gv) and environment (Ev), to scaffold learning of genetics. In short, we want to examine how students think about and understand Pv = Gv + Ev.
As a starting point for curricular assessment we asked the education research question: To what extent does an integrative, inquiry-based genetics unit influence students’ cognitive structures and explanations of phenotypic variation as a quantitative trait? We hypothesized 1) that students’ cognitive structures in the form of word associations to the stimulus word “genetics” would shift and reveal “variation” as a prevalent concept after the unit and 2) that students would be able to articulate accurate and complete explanations of inheritance and expression of phenotypic variation using reasoning that integrates understanding of variation in genotype and environment. Our approach examined students’ cognitive structures and familiarity with concepts through free word association and self-reported understanding of particular genetics and evolution concepts on a pre- and postunit survey. We then examined students’ explanations through analysis of their final scientific research posters and coded their explanations for phenotypic variation of anthocyanin pigmentation as a quantitative trait that is influenced by genotypic variation and the environment. Bahar et al. (1999; and previously, in Shavelson, 1972) suggests that retrieval of words and concepts from long-term memory through word association reflects the cognitive structure for how students relate concepts. Free word association is a common method to examine students’ cognitive structures (Shavelson, 1972; Bahar et al., 1999; Tsai and Huang, 2002) and has gained use as a research tool for evaluating semantic networks and retrieval of related conceptual knowledge (DeDeyne and Storms, 2008; Steyvers and Tenenbaum, 2005). Word association is a particularly powerful tool for evaluating understanding when interpreted together with written explanations, concept maps, drawings, or interviews (Bahar and Hansell, 2000; Hovardas and Korfiatis, 2006; Kurt et al., 2013).
METHODS
Course Context and Student Participants
This study took place in Fall 2011 and 2012 with 107 and 102 student participants, respectively, enrolled in an introductory Ecology, Genetics, and Evolution lab course. This is the foundational lab course in Biology Core Curriculum (Biocore), the four-semester honors biology program at University of Wisconsin–Madison (described further in Batzli, 2005). The course goals emphasize the development of process of science skills, integrative learning, group-based learning, and scientific communication through a progression of multiweek inquiry-based units. The unit described here is the second of three units in the 15-wk course. Along with the learning outcomes stated above, major learning goals and materials for this lab unit are outlined in a custom lab manual chapter (see Supplemental Material 1). Students participating in this study were enrolled in one of five lab sections of ∼22 students each. Each lab section met weekly for a 50-min discussion period followed by a 3-h lab period, with the discussion and lab separated by ∼24 h. All students enrolled in the course signed informed consent to participate in the study before the beginning of the lab unit (approved IRB 2008-0783). Students in the study population were 49% male and 51% female; 83% had declared majors including genetics (17%), biochemistry (40%), and biology (28%); 51% had taken the Advanced Placement (AP) Biology exam, scoring 4 or 5; and their average cumulative grade point average was 3.17. All students had previously or were concurrently taking the companion lecture course (Ecology, Genetics, and Evolution) in which the genetics unit was taught using a combination of lecture and in-class problem-solving activities that emphasized meiosis, inheritance of Mendelian traits, and genetic analysis (e.g., monohybrid, dihybrid, linkage, three-point cross). Quantitative genetics was covered in one 50-min class meeting that included a minilecture interspersed with clicker questions and one in-class problem-solving exercise. The material emphasized in class was supported by assigned textbook readings and an online narrated set of PowerPoint slides emphasizing concepts and calculations of narrow sense heritability, selection differential, and response to selection. Although most of the basic concepts introduced in lecture and textbook readings were applied in lab, there was no explicit emphasis on variation or on Pv = Gv + Ev in the lecture course.
Curriculum and Unit Design
Before beginning the unit, students completed a required prelab assignment (Supplemental Material 2) with questions and problems that could be answered based on careful reading of the lab manual chapter as well as previous instruction on elementary statistics and how to generate descriptive statistics and frequency histograms using Microsoft Excel.
In week 1 of the unit, students were introduced to the driving question “What determines phenotype (Pv = Gv + Ev)?” As we began, we drew from students’ prior knowledge of genetics with acknowledgement of everyday vernacular use of concepts such as gene, genetic variation, and expression. The unit instructors (J.M.B., S.A.M., and P.H.W.) then guided students through an introductory dialogue using Socratic questioning and a set of PowerPoint slides (Supplemental Material 4) in which students’ pre-existing vocabulary was used in new situations to discuss families, genetic relationships (identical twins, fraternal twins, siblings, half-siblings), and when/how variation in phenotype arises. We assigned students to research teams of four to begin work on a concept-mapping exercise that explored the concepts of genes, genetic variation, alleles, environment, phenotype, genotype, discrete and quantitative traits, artificial and natural selection, and the nature of phenotypic variation in humans and other animal and plant species. The class then transitioned to a discussion introducing Brassica spp., RCBr genetics, and plant reproductive biology. Finally, each team of four students made observations of 1-wk-old Fast Plants seedlings from P1 (green = homozygous recessive for the Mendelian inherited anl− allele), P2 (purple = homozygous dominant), and F1 (purple = heterozygote) populations, and measured the intensity of anthocyanin expression in the hypocotyl region for six plants in each population using a custom-made anthocyanin pigment color index tool in which color ranged from 0 = green to 4 = dark purple pigmentation.2,3 Students entered their data into a common spreadsheet that included combined data for all lab sections and discussed patterns of phenotypic variation they observed in the data. At the conclusion of the first lab period, we asked students to predict the average anthocyanin pigmentation of a selected F2 population (F2 selected or F2s) generated from interbreeding the most highly pigmented individuals in the F1 population (interbreed 25% most highly pigmented, i.e., deepest purple) as compared with 25% randomly interbred set of individuals (F2 drift or F2d). We also asked students to brainstorm environmental conditions that might alter the expression of the anthocyanin trait. These two predictions, 1) the influence of genotype (F2s compared with F2d) and 2) the influence of the environment on phenotypic variation of F2s and F2d populations, became the foundation of the students’ hypotheses and research proposals that they presented to their lab section the following week. We assigned an experimental design worksheet to guide student research teams in their background research, development of rationale for hypotheses, and design of their experiments (Supplemental Material 5).
In the second week of the unit, student teams prepared a short (15 min) PowerPoint presentation of their research proposals to solicit feedback from the class. Teams presented specific hypotheses and rationale with supporting literature, described their methods, explained their expected and alternative results in graphical form, and described potential implications of their research. Through peer-to-peer feedback and instructor comments, the research teams refined their experiments. In addition to research proposal presentations, students cross-pollinated F1 individuals within the 25% most highly pigmented group (n = 25% of the population) and then cross-pollinated an equal-sized population of randomly selected F1 individuals, as done in Goldman (1999). Pollinated F1 plants were left to set seed for the production of F2s and F2d populations, respectively, used in the subsequent semester. Given the length of time it takes for one generation to produce seed (40 d), the F2 seed stock used by students in this study was produced in the summer preceding the lab unit. Student teams planted seeds for their investigations outside of class time and placed their growing containers in both control (see optimal growing conditions listed in Appendix 3 of Supplemental Material 1) and experimental conditions as guided by their research proposal. Experimental conditions included alterations of light (quality or quantity), UVB exposure, changes in nutrients (quality and quantity) or pH, addition of sucrose, simulated herbivory, phototropism, gravitropism, changes in temperature or humidity, simulated drought or flooding, or simulated wind exposure.
We describe remaining activities that defined the core of the 4-wk curriculum in Supplemental Material 3. As the final assignment for this unit, each student generated a scientific poster describing his or her experiment; his or her rationale, hypothesis, and research methods; and an explanation of the data as evidence for phenotypic variation in anthocyanin pigment intensity as influenced by Gv and Ev. Our expectations for poster development and for explanations were detailed as guiding questions and were made available to students at the beginning of the unit (see Supplemental Material 6). We provided this guidance to help students structure their thinking and explanations in their posters.
Throughout the unit, all instructors and teaching assistants made every effort to model accurate and precise language when using terms such as variation, gene, allele, genetic variation, phenotype, genotype, adaptation, recessive and dominant, organism, individual, and population, since incorrect and/or inconsistent usage of these terms has been associated with known misconceptions (Anderson et al., 2002; Smith et al., 2008) or inaccurate understanding (Bizzo and Caravita, 2012).
Assessment Data Collection
We asked students to complete a short online survey 2 wk before the start of the unit (n = 98 and 95 respondents in 2011 and 2012, respectively) and again 2 wk after the conclusion of the unit (n = 95 and 102, respectively). In the survey, we asked students to conjure words or concepts in association with the single stimulus word “genetics” (i.e., “List the first 5 concepts that come to mind when you think of genetics”). We choose “genetics” as a stimulus, rather than “variation,” to gather a broad census of students’ cognitive structures from their prior knowledge going into the unit and from their knowledge of genetics after the unit. Students reported each concept as a single word (e.g., chromosome) or phrase (e.g., independent assortment, Punnett square) separated by commas. In addition, we asked students to rate, on a 3-point Likert scale (never heard/don't understand, intro to intermediate understanding, advanced understanding), their understanding of common words associated with genetics and evolution that would be used in the unit (i.e., phenotype, phenotypic variation, genotype, gene, genetic variation, allele, discrete trait, quantitative trait, heritability, phenotypic plasticity) to measure students’ self-reported familiarity with the concepts. Although demographic questions (i.e., gender, major, prior genetics courses) were included in the survey, we did not ask students to provide their names or any other identifier (e.g., student identification number). Finally, in 2012, we collected students’ final scientific posters and randomly sampled 53 from a total of 102 across all five lab sections to examine student explanations of the influence of variation in genotype and environment on the inheritance and expression of anthocyanin pigmentation using a rubric developed to analyze students’ understanding of Gv, Ev, and Pv (Table 1).
| Influence of genotypic variation (Gv) | Influence of environmental variation (Ev) and Gv × Ev interaction |
|---|---|
| 1. Quantitative trait is conditioned by multiple genes expressed through biosynthetic pathway. | 1. Quantitative trait is plastic to the environment. |
| 2. Trait is heritable with selected offspring having higher anthocyanin intensity than parents or drift population. | 2. Mechanism for how environment influences expression or alteration of phenotype. |
| 3. Selected population has higher anthocyanin intensity than other populations conferred by change in allele frequency. | 3. Reasoning for how environment influences genotypically unique populations. |
| Level of explanationb | |
| Missing Completely missing or exceptionally vague | |
| Low Missing two or more elements; two elements present but vague, inaccurate, or incorrect explanation | |
| Intermediate All elements present but vague or some inaccuracy; two elements present and accurate | |
| High All elements fully present and accurate | |
Assessment Data Analysis
Free Word Association.
We analyzed students’ pre–post responses to the word association question in aggregate and in three ways: 1) overall word species frequency, 2) word diversity, and 3) paired word co-occurrence matrices. The purpose of the word association was not to evaluate student responses as either correct or incorrect but rather to examine students’ cognitive structures in aggregate as they are associated with genetics and how relationships between concepts change through exposure to this rich type of integrated, inquiry-based investigation. We assumed the word association task to be of low cognitive load and that retrieval of concepts in association with “genetics” was an instantaneous snapshot of a student's close cognitive connections to the stimulus (Shavelson, 1972; Bahar et al., 1999). Because students reported their word association in a string of comma-delineated text, we assumed that each response word or phrase was not only associated with “genetics” but also with the other response words in the chain.
We generated word counts and frequencies of the composite set of all concepts and phrases conjured by the entire student population regardless of the order in which individual students reported the words. Word counts were used to determine the most frequent concepts the student population associated with genetics in pre- and postunit surveys across 2011 and 2012. Frequency was calculated by dividing the number of occurrences of each single word (e.g., variation) or multiword phrase (e.g., Punnett squares, gene by environment interaction) by the total number of students’ responding. Frequencies of 0.05 or higher in the pre- or postunit survey were further analyzed to calculate the degree to which the word frequencies had changed pre- to postunit.
We analyzed word diversity on dimensions of word richness (total number of different words) and evenness (the extent to which the words appear in equal frequency) using a Shannon diversity index. The Shannon diversity index is often used in ecological science to describe biological species diversity of a particular habitat (Shannon, 1948). In this case, we define word diversity index as 
We then assessed students’ five-word clusters for patterns of co-occurring words within the population where more than 20 instances of a word pair appeared within clusters. We did this by generating a matrix representing counts for each occurrence of unique word pairings within student responses for each year and pre/postunit grouping. Word-pairing counts were standardized by total number of student respondents and were relativized to a scale of zero to one (high relatedness) by dividing all cells by the maximum standardized relatedness score across years and pre/postunit group. We then visualized relatedness scores colorimetrically for word pairs that co-occurred within pre/postunit groupings and between years.
Word frequencies and word occurrence/co-occurrence matrices were made in R statistical software version 3.0.2. Data matrix visualizations were made using the corrplot library and were modified in Adobe Illustrator for aesthetics.
Examination of Poster Explanations.
We analyzed students’ final research posters following the conclusion of the 2012 semester. All posters were deidentified before analysis, with student names and section numbers removed. Ten posters were chosen randomly to help in the construction of a rubric with two subdimensions: 1) explanations on the influence of genotypic variation (Gv) on phenotypic variation and 2) explanations on the influence of environmental variation (Ev) on phenotypic variation. We separated the subdimensions of Gv and Ev because we recognized that students often talk about and may learn about genetic and environmental influences as separate noninteracting entities (e.g., nature vs. nurture). That said, many student research teams observed Gv × Ev interaction happening in their experiments. Therefore, we also looked specifically for Gv × Ev explanations in their posters as one element of Ev explanations. Concepts evaluated by the rubric addressed learning outcomes described above and were the focus of the majority of instructional materials and class discussions. The rubric is detailed in Table 1 and includes elements for each subdimension and levels coded for explanations ranging from high level to missing. Each poster was reviewed thoroughly and text from introduction and discussion sections was excerpted for coding. Two independent raters (J.M.B. and K.D.) scored 53 posters using the rubric in two iterations with refinement of the rubric in between resulting in an interrater score of 81 and 69% agreement (Cohen's kappa 0.71 and 0.55) on the Ev and Gv subdimensions, respectively. We analyzed the relationship between subdimensions by student using a Spearman's rho (ρ) nonparametric correlation test.
RESULTS
Free Word Association
Frequencies of words and phrases grouped by year and pre- and postunit survey are reported in Table 2. In both 2011 and 2012, the words “meiosis,” “mitosis,” “chromosome,” and “DNA” were mentioned at high rates in preunit survey and then attenuated in the postunit survey, while “allele,” “genotype,” “selection,” and “variation” show the opposite pattern. Other words emerged in the postunit word associations that were not present in the preunit, including “phenotypic plasticity.” Words that did not change in frequency between pre and post in either year included “mutation” and “Mendel.” The word “heritability” was prevalent in the postunit survey in 2011, but did not appear to any great extent in 2012.
Through analysis of aggregate frequency change for the most prominent words (> 0.05 frequency) included in student word associations in 2011 and 2012 (Figure 1), we found a shift in pre- (right side of y-axis) to postunit (left side of y-axis) word association frequency. We observed the following set of concepts with higher prevalence (positive change in frequency) in the preunit survey across both 2011 and 2012 (indicated with solid orange lines): “mitosis,” “meiosis,” “DNA,” “replication,” “chromosome,” “Punnett squares,” “genetic disease,” and “mutation.” In comparison, the set of concepts most prevalent in the postunit word association (negative change in frequency) across both years (indicated with solid black lines) included “recombination,” “Mendelian genetics,” “traits,” “dominant,” “genotype,” “selection,” “phenotypic plasticity,” “evolution,” “allele,” “variation,” and “heritability.” We observed only four prominent words that switched in prevalence between 2011 and 2012 (indicated with dashed lines): “inheritance,” “gene,” “Mendel,” and “phenotype.”
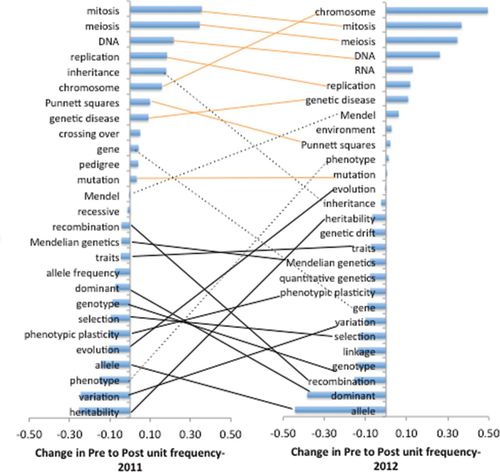
Figure 1. Aggregate word frequency change from preunit to postunit word association task given the stimulus word “genetics” in 2011 and 2012. Lines connect identical words as they appear in 2011 and 2012, with orange lines indicating positive change in both years, black lines indicating a negative change in both years, and dashed lines indicating a switch between years. (Pre- and postunit: n = 98 and 95 in 2011; n = 95 and 102 in 2012.)
The relatedness matrices illustrated in Figure 2 highlight occurrence and co-occurrence of the concepts in five-word clusters that students associated with genetics in 2011. The upper-left to lower-right diagonal in the matrices colored in shades of red represents relative frequency or occurrence of each word. Paired word co-occurrences are colored in shades of blue (light blue = low co-occurrence, dark blue = high co-occurrence). As with the frequency-change analysis (Figure 1), Figure 2 reveals a clear shift in word occurrence and co-occurrences in student word clusters between pre- and postunit surveys. In the preunit matrix, areas of brightest blue indicate high frequency of co-occurring concepts of mitosis, meiosis, chromosome, gene, and DNA. Other high-frequency co-occurring concepts colored with bright blue are allele, heredity, mutation, and Punnett squares, which co-occur with each other and with the concepts listed above. Intermediate-frequency co-occurring concepts of phenotype and genotype, shaded in intermediate blue, co-occur primarily with each other but also with DNA. The word “trait” occurs at a similar frequency similar to phenotype and also, like phenotype, co-occurred most frequently with DNA. The words “mutation” and “gene” co-occurred consistently in pre- and postunit word associations, with a similar pattern in both years. Interestingly, Mendel occurred at intermediate frequency, but did not co-occur with any particular concept. In the postunit matrix, the patterns of co-occurrence noted above become disaggregated. The bright blue “hot spots” surrounding meiosis, mitosis, DNA, and chromosome soften to intermediate shades indicative of evenness in relative frequencies among concepts. The set of concepts represented at intermediate to high co-occurrence in the postunit matrix are allele, chromosome, DNA, gene, genotype, mutation, phenotype, heritability, and variation.
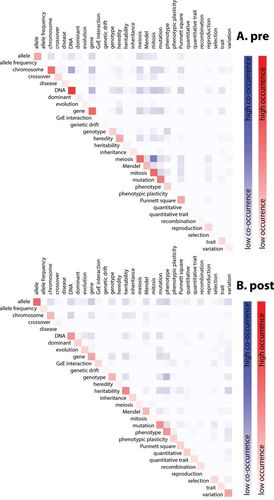
Figure 2. Word matrices for 2011 pre- and postunit survey of student five-word cluster word associations representing words that occurred in at least 20 student responses across both years (see Supplemental Material 7 for 2012 matrices). Each red shaded cell on the diagonal represents individual word occurrence or relative frequency. Each blue cell colorimetrically represents paired word co-occurrence (dark blue for high co-occurrence, light blue for low co-occurrence). All cell scores were standardized by total student respondents and were relativized to a scale of zero to one (high relatedness) by dividing all cells by the maximum standardized frequency of occurrence or co-occurrence across years and pre/postunit groups. (A) Preunit occurrence and co-occurrences (n = 98); (B) postunit occurrence and co-occurrences (n = 95).
When probing the matrix for the proposed threshold concept “variation” in the preunit matrix, we found it in low frequency and observed no pattern of co-occurrence with other concepts. In postunit analysis, variation was found to co-occur at high relative frequency with allele, heritability, and phenotype. Owing to space limitations, we present the co-occurrence matrices for 2012 in Supplemental Material 7. Similar patterns of hot spots and co-occurrences were observed between years with only minor differences, all except for the notable change in co-occurrence of variation with mutation and not with heritability. Even though “variation” did not co-occur with heritability, both heritability and variation co-occurred with mutation at intermediate frequency.
Through analysis of word diversity, we found students associated 105 (pre) and 104 (post) different words or phrases with genetics in 2011. In 2012, the number of pre to post words totaled 106 and 95, respectively. There were 44 and 42 words found in common between the 2011 and 2012 pre- and postunit word analyses. We calculated preunit word diversity indices of 4.73 and 4.41 and postunit indices of 5.23 and 4.82 in 2011 and 2012, respectively. When we analyzed word frequency and word diversity together, there were approximately the same number of words but a higher diversity of words in the postunit as compared with the preunit, indicating a greater evenness or greater consistency in words and phrases revealed through word association among our student population at the end of the unit.
From surveys of self-reported understanding before beginning the unit, few students had heard of or had familiarity with the concepts of artificial selection, genetic drift, independent assortment, segregation, discrete trait, phenotypic plasticity, or quantitative trait, with 25–62% of students reporting that they had never heard of or do not understand these concepts (Figure 3). This is in contrast to the other common genetics terms that students had intermediate to advanced self-reported understanding of in the preunit survey. By the end of the unit in the postunit survey, the vast majority of students (81–98%) reported at least intermediate-level understanding of concepts, including those they were not familiar with before the unit. Important to our goals, 55–58% of students reported advanced-level understanding of genetic variation and phenotypic variation.
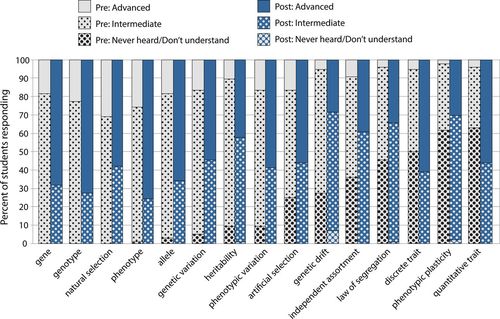
Figure 3. Self-reported student understanding of genetics terms on the pre- and postunit survey (n = 98 and 95). Data reported here include the percent of students reporting advanced understanding (working knowledge or deep-level understanding), introductory to intermediate understanding, or never heard/don't understand.
Student Explanations
Our analysis of student posters revealed a normal distribution in levels of understanding of student explanations on both subdimensions (Ev and Gv; Figure 4). We found no apparent correlation between students’ individual explanations of the Gv and Ev subdimensions (Spearman's ρ = −0.02, p = 0.88). Of the 53 posters scored using the rubric detailed in Table 1, 51% of students articulated intermediate- to high-level explanations for how Ev would influence phenotype, with high-level explanations including discussion or speculations for how and why the environment would influence genotypically distinct populations (F2s and F2d) differently or somehow alter the phenotype (chemical alteration of anthocyanin pigment; see examples in Table 3). Similarly, one-half of students explained the influence of genetics at an intermediate to high level, including reasoning for how the quantitative trait anthocyanin pigmentation is conditioned by multiple genes and that heritability of a quantitative trait can be measured using artificial selection.
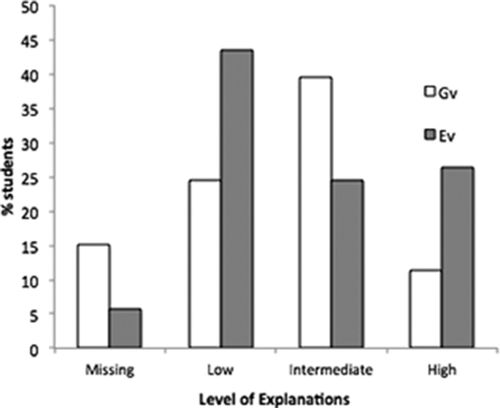
Figure 4. Percent of students’ explanations on the influence of genotypic variation (Gv) and environmental variation (Ev) at each rubric level (missing, low, intermediate, and high) through analysis of final research posters (n = 53).
| Explanation | Level | Example quotations from student final research posters. |
|---|---|---|
| Influence of genotypic variation (Gv) | High | “Anthocyanin is produced through the flavonoid biosynthesis pathway, which involves many gene loci suggesting that intensity of pigment is a quantitative trait. Plants with more intensely purple stems contain a higher proportion of alleles for high anthocyanin production and selection can increase these allelic frequencies in offspring.” |
| Intermediate | “Intensity of anthocyanin pigmentation is controlled by multiple genes [coding for] biochemical intermediates, and it is heritable as a quantitative trait. Offspring show similar phenotypes [to] their parents in a common environment through artificial selection.” | |
| Low | “Anthocyanin is a quantitative trait where each gene involved in the biochemical pathway contributes a ‘dose’ of anthocyanin. When many genes influence a phenotype like anthocyanin in B. rapa, selection for purple genes creates purple offspring.” | |
| “[Anthocyanin is] influenced by number of doses of dominant alleles. If Fl is bred for the ANL allele, then offspring will have an increase in ANL alleles (F2s).” | ||
| Influence of environmental variation (Ev) | High | “Exposure to increased UVB [light] up regulates gene expression to aid in anthocyanin production.… significantly greater increase in F2s [pigmentation] as compared with F2d in UVB treatments was most likely due to F2s populations having a greater potential for expression due to selection in Fl for higher expressing alleles.” |
| Intermediate | “Sucrose is the major form of soluble C for long-distance transport in plants. [Sucrose is] shown to up regulate anthocyanin-related genes by acting as a signal molecule and creating a C-surplus environment, stimulating Shikimate pathway to produce anthocyanin.” | |
| Low | “B. rapa has strong expression of anthocyanin in abundant light; anthocyanin production is less intense in reduced light. Ornamental grasses (similar to B. rapa) are purple in abundant light and pale violet/green in low-light environments.” |
Although relatively rare, high-level explanations of genetic influence revealed sophisticated understanding of anthocyanin pigmentation as a quantitative trait and how artificial selection leads to inheritance of alleles with high potential for expression of anthocyanin under specific environmental conditions (Table 3). There were a number of intermediate-level explanations for Gv that were missing a description of how selection and drift lead to a predicted change in allele frequency and higher frequency of deeply pigmented plants in the F2s population. Most low-level explanations of genetic influence (36%) were missing key elements and included fully or partially inaccurate, incomplete, and/or missing explanations. One concept that was unexpectedly and inaccurately applied in student explanations was “gene dose.” Three students in our sample explained the difference between low- and high-intensity pigmented plants, saying that low-intensity pigmentation resulted from plants inheriting fewer genes involved in anthocyanin biosynthesis, whereas high-intensity pigmented plants inherited more genes, resulting in a higher “gene dose.” Students that invoked “gene dose” explanations did not discuss or infer polyploidy or aneuploidy. In addition, a few students (3 of 53) attempted to explain phenotypic variation of the quantitative trait using logic and reasoning of a Mendelian trait describing highly pigmented plants as those in the homozygous dominant condition and higher frequency of homozygous dominant plants in the artificially selected F2s population. Other observations included the misuse of the words “allele” and “gene,” conflating the terms or using them inaccurately when inferring gene loci or the heterozygous or homozygous, recessive or dominant condition. Given the prominence of the word “allele” and “gene” in students’ postunit word associations, we looked for evidence of correct versus incorrect use of the terms in Gv explanations of anthocyanin inheritance and expression. We found students misused the word “allele” in five of the 33 (15%) instances where it appeared. In addition, there were several examples wherein students confused or had limited capacity to explain the genetic variation (or lack thereof) underlying genetic drift and phenotypic plasticity or incorrectly explained phenotypic plasticity as the result of artificially selecting for more genes associated with the trait.
Although we did not require students to fully explain the mechanism of the discrete trait included as a “control” or Mendelian marker for the breeding schema, the majority of students were able to confirm and provide accurate explanations in their posters that the inheritance of the green plants in F2 populations was the result of segregating the anthocyaninless allele in a Mendelian 3:1 ratio. Students discussed this ratio as a control or check that the crosses and pollination were done correctly with minimal contamination.
DISCUSSION
Word Associations and Cognitive Structure
We found that students’ word associations with genetics shifted as a class population through this unit with different word associations emerging from pre- to postunit surveys. From the comparison of pre- and postunit word frequency (Table 2) and frequency-change analysis (Figure 1), it is clear that students come to the course with prior (preunit) knowledge of terms and concepts typical of high school genetics curricula, including concepts introduced in AP Biology (https://apstudent.collegeboard.org/apcourse/ap-biology/course-details), which 51% of the students had completed. By the end of the unit, the word frequency-change analysis revealed many of the concepts emphasized through instruction and students’ inquiry as a part of this unit. We noticed a surprising consistency between years 2011 and 2012, suggesting that the shift in student word associations as a class is repeatable and responsive to this curriculum. Greater word diversity combined with a shift in word frequency in the postunit word association is likely the result of language learning together with some language conformity upon completion of the unit. For instance, the words “phenotypic plasticity” and “variation” appear at intermediate frequency in students’ word associations at the end of the unit, but appear seldom or not at all in the preunit word associations (Table 2). Most students (61–63%) self-reported that they had “never heard/don't understand” the terms phenotypic plasticity and quantitative trait in the preunit survey, but they reported intermediate- to advanced-level understanding in the postunit survey (Figure 3). Regardless of students’ understanding of the new terms, the pre- to postunit shift in word association illustrated through change in frequency (Figure 1) and incidence of co-occurring concepts that are new to students through this unit suggests that students are integrating new pathways for learning genetics into their cognitive structure (Bahar et al., 1999; Tsai and Huang, 2002).
Taken as a class population, shifts in word frequency (Figure 1) and in word-clustering relatedness matrices (Figure 2) are indicative of changes in the class's cognitive structure as they progress through the curriculum. The shift in students’ association from foundational concepts such as mitosis, meiosis, DNA, and chromosome to more discipline-specific concepts of phenotype, genotype, and allele together with the proposed threshold concept of variation may be an indication of students’ development of integrative understanding within the discipline. Taylor (2006) discusses how threshold concepts in biology are abstract and difficult to understand unless they are used in the process of biology, as variation was applied in this inquiry-based unit. As a holistic concept, “variation” in all its forms (e.g., genetic, allelic, heritable, environmental, genotypic, and phenotypic variation) has the potential to serve as a gateway and integrative concept—a key concept leading to higher-level understanding of genetics and evolution (Ross et al., 2010; Duncan and Rivet, 2013).
Our data from word associations allow only a preliminary interpretation and require much greater study about how “variation” as a concept and process, as well as other proposed threshold concepts of randomness and scale, may serve as major nodes for formulating learning goals in genetics and evolution curriculum. We found word association to be a valuable research tool when paired with relatedness/co-occurrence matrices that can be probed for particular concepts to visualize primary, secondary, and even tertiary connections. They are easy to administer, of low cognitive load for students, and informative for queries into instructional practices.
Student Explanations
We used the analysis of student explanations in final posters as a measure of student understanding. While we recognize that the poster format may have confined students’ depth of explanations, approximately half of the students were able to produce intermediate- to high-level explanations for phenotypic variation as a function of variation in genotypic variation and environmental variation (the main learning goal of the unit; Figure 4). Students cited many papers in the primary literature to support their environmental explanations and were able to connect their understanding of environmental stimuli as signals for up-regulating or down-regulating key elements in anthocyanin biosynthesis. Although none of the students had taken a college-level course in molecular biology or biochemistry, the learner-centered basis of their inquiry may have inspired deeper learning of how environmental stimuli can influence phenotype at the molecular/biochemical level.
While half of the students demonstrated high-level explanations, the other half of the class revealed low-level, inaccurate, or incomplete explanations with some important misconceptions uncovered (e.g., gene dosage). The example of gene dosage is an interesting case of what seems like a misplaced formulaic phrase students used to try to explain a genetic phenomenon about which they had limited conceptual knowledge. In this case, it seems that students were utilizing vernacular understanding of the concept “dose” and assigning it scientific merit by preceding it with the concept “gene.” This is a common strategy used in constructing understanding, one in which students backslide into an interlanguage that is part vernacular and part scientific (Olander and Ingerman, 2011; Rincke, 2011). Although these students indicated partial understanding, that multiple genetic elements were involved in regulating the amount and intensity of anthocyanin produced, it seemed they were conflating the concept of gene and allele. In fact, the most prevalent word revealed in the postunit word association surveys was “allele” (Table 2), which also was the word most frequently misused in poster explanations, often used interchangeably with “gene” (9% of the time).
Although we recognize that self-reported understanding does not reconcile well with direct measures of student understanding (Falchikov and Boud, 1989; Willingham, 2003/2004), the differences in word associations in the preunit survey compared with the postunit survey is suggestive of gains in language acquisition from which to start formulating understanding. Sherin et al. (2012) describe more generally how students construct scientific explanations from conceptual nodes in their cognitive structure to form “dynamic mental constructs” that shift and converge on a formed explanation through interviews and probing. Ideally, we would like to align students’ word associations and explanations with an interview to allow students a more flexible forum to describe their thinking on how Pv is influenced by Gv and Ev in their experiments. We predict that this approach could reveal a more thorough view of their conceptual understanding.
One issue that may have weakened students’ explanations was if they were unable to measure a difference in phenotype between F2s and F2d in their experiment. If research teams did not observe a gain from selection, their explanations moved in the direction of random-sampling error or limits in their capacity to detect differences in one generation. Although their interpretations were appropriate, their explanations lacked evidence of understanding of the genetic principles they were testing in their experiments. Therefore, it is possible that some students could explain phenotypic variation well but did not have the opportunity to do so given the data they generated. On the other hand, students may not have gained a thorough exposure to the breadth of genetics there was to explore in this unit if their data were difficult to interpret or counterintuitive. Duncan and Rivet (2013) discuss science learning as a progression that requires an intermediate and developmentally appropriate stage during which students entertain inaccurate “yet productive” explanations that then lead to higher-level understanding. If students are able to detect biologically relevant results through their experiments, they have an opportunity to construct productive explanations. Otherwise, their explanations may be inaccurate, making the learning unproductive. This is not a call for greater structure of or guidance in the inquiry and student experiments such that the results become confirmatory, but rather a call to pay close attention to each team's data and provide opportunities for comparison or for pooling class data to look at general patterns in the data that could inform student explanations in their research posters.
Interestingly, there was no correlation between students’ Gv and Ev explanations. Student explanations for how genotype influences phenotype seemed disconnected and independent of their explanations for how environment influences phenotype. As this was an inquiry-based lab unit, students were permitted to choose an environmental treatment they predicted would influence expression of anthocyanin intensity, and they dove deeply into literature to support their hypotheses for how the environment may influence phenotypic variation (for review papers, see Holton and Cornish, 1995; Lee and Gould, 2002). They did not, however, get to choose the phenotype to study, nor did they create the breeding and selection scheme that provided the genotypic backdrop for the plant populations they studied. Student learning about the environmental influence was more self-directed and may have seemed quite separate from doing the genetic crosses and learning about the genetic background of the plant populations. Unless students were prompted to discuss and explain why F2s and F2d responded differently or in an additive way to the environmental treatment based on the data they collected at the end of the experiment, they kept their Gv and Ev explanations separate. Once we recognized this challenge, we focused on discussing gene by environment (G × E) interaction during week 3 and 4 of the unit in the subsequent year's iteration of the unit in 2013. Gene by environment interaction is a difficult concept but very interesting for students to explore. G × E interaction represents the intersection of much current research (Grishkevich and Yanai, 2013) and provides an excellent launchpad for learning 21st-century genetics (Hunter, 2005; Plomin et al., 2009).
Implications for Instruction
Our results suggest that the content or approach to learning in this integrative genetics unit is challenging and requires great attention from instructors as to what students are saying and how they are explaining their observations. Class discussions, feedback presentations during weeks 1 and 2, and feedback loops during preliminary and final data collection (weeks 3 and 4) are essential curricular components for building students’ cognitive structures and for determining students’ understanding and readiness for moving on to more complex concepts such as G × E interaction.
Some challenges to learning led to productive instructional breakthroughs. An example appeared at the beginning of the unit (week 1), when few students could separate the notion of dominance (purple) and recessiveness (green) of the discrete trait (Mendelian marker) from the continuous gradation in anthocyanin pigment intensity as the quantitative trait. Many students confused the phenotypic “blending effect” of incomplete dominance at a single gene locus with the range of pigmentation intensity revealed in their B. rapa populations. Anticipating this learning challenge, we helped students use their understanding of Mendelian genetics to make tentative predictions of phenotypic variation for F2s and F2d populations based on measurement and analysis of parent and F1 populations during week 1. Through their analysis and a discussion, we converged on the question “Why do F1 individuals vary in anthocyanin intensity if all plants are heterozygous (Aa) for the anthocyaninless (anl−) allele?” This question led to the metaphor of discrete traits being controlled by single gene “on/off” switches. When the switch is “off,” as it is for homozygous recessive individuals, then plants are green. When the switch is “on,” as it is for all the F1 heterozygous individuals, then a panel of many “rheostat” levers or “dimmer” dials take over and control for the level of intensity. The “dimmer” dials, all set at different levels, represent the multiple genes and many allelic variations involved in conditioning expression of anthocyanin pigmentation in a gradual way from very low to high pigmentation. Using this metaphor of on/off switches and dimmers, we discussed the Mendelian trait, anthocyaninless, as a type of genetic “control” or Mendelian marker trait to help move the conversation beyond Punnett squares into multigenic inheritance and expression. Interestingly, we found very few students who confused the concepts of Mendelian genetics and quantitative genetics by the end of the unit as evidenced in their final posters explanations.
One challenge that we did not anticipate was the considerable difficulty students had in understanding how parents and offspring (P1, P2, F1, and F2 populations) plant populations could be grown at the same time. This obstacle to understanding could have contributed to some students’ limited capacity to explain how the F1 population is fundamentally related to the F2 populations and how alleles from the F1 are represented in F2. Furthermore, 25–30% of students reported that they had never heard of artificial selection or genetic drift before the unit (Figure 3). A more thorough discussion of plant reproduction, including seed dormancy and cross-pollination, during week 1 of the unit may help students better recognize the genetic relatedness and pedigree of their populations. Although we intentionally delayed an in-depth discussion of these powerful mechanisms of evolution to shift allele frequencies, in hindsight, we could have introduced these concepts earlier in an activity or simulation to better prepare students conceptually for the lab. The consequence of students misunderstanding the underlying breeding scheme, the relatedness of the populations, or the concepts of selection and drift may have resulted in students’ lack of ability or missing explanations on the influence of genetics on phenotypic variation in their posters (Figure 4).
For most students, this was the first time they had considered the molecular scale of specific genes and proteins being associated with a very visible and measurable macroscale trait such as anthocyanin pigment intensity. Although it was challenging to translate their basic understanding of the central dogma from high school biology to this context, many students had aha moments while in the process of making predictions or interpreting their results. We believe that the rich process of inquiry, development of testable questions, refining of hypotheses, discussion of biological rationale for hypotheses, and use of data as evidence made these aha moments possible.
We emphasized “variation” as the primary learning goal for the unit—and specifically probed the word association matrix for it. Given the co-occurrence of “heritability” with “variation” in the 2011 postunit word association matrix, we assume that students somehow relate the two concepts. It may be that wrestling with the idea and calculation of narrow sense heritability allows students to view variation in a more integrated, applied, or relevant way. Interestingly, “heritability” did not co-occur with variation in the 2012 matrix (Supplemental Material 7). It is worth noting here that we expected students to calculate narrow sense heritability estimates as a part of their final project in 2011 but removed this expectation in 2012, since students tended to misinterpret the meaning of heritability estimates (e.g., when students calculated low heritability, they often concluded that there was no genetic basis for variation in the trait). With the findings from this analysis, we are reconsidering how we might return “heritability” estimation and interpretation to the curriculum in future years and add in discussion of epigenetic interactions.
This unit provided many opportunities for students to practice, refine, and integrate their understanding of concepts in genetics, quantitative genetics, and Mendelian genetics with evolution through inquiry learning and the process of science. White et al. (2013) present another recent example of an integrative genetics and evolution curriculum, but instead of weaving in quantitative genetics, they utilize Mendelian genetics only in a case-based approach to integrate genetics and evolution of traits in a nonlaboratory setting. We believe there is great value in working with a quantitative trait in combination with a Mendelian trait, therefore allowing students to explore and ask questions about variation and resolve meaningful patterns, which they could not do if they were working with Mendelian traits alone. In our experience, when the curriculum is limited to Mendelian genetics, we observe students falling into a pattern of binary thinking that obscures their ability to reason about variation they see beyond what can be explained with Punnett squares. When students combine reasoning from Mendelian and quantitative genetics, they expand their toolbox and think about the real and multigenic dynamics of more than one loci being involved in the phenotypic variation they observe. We believe that it is important to decrease students’ reliance on the patterns and expected ratios of Mendelian genetics to solve all genetic problems and to provide rich opportunities to explore gene expression, epigenetics, environmental stimuli, and signal transduction among other topics. Although the curriculum of White et al. (2013) does not introduce quantitative genetics, it emphasizes epistemic goals similar to the ones presented here. Therefore, the two curricula may be well paired as a case-based “lecture” and “inquiry-based” lab.
Recommendations for Implementation
Although there are demonstrable advantages to this approach to learning genetics, this unit is not “plug and play.”
Begin with prior knowledge: survey students’ background knowledge and comfort level with genetics to help scaffold the type of language and guidance students may need at different points in the 4-wk curriculum. For instance, introducing the unit as “quantitative genetics” may make it seem hard or foreign to students and, therefore, it may be wise to avoid using this phrase until later in the unit, when students’ cognitive structures have progressed or are more developed. Rather, refer to anthocyanin pigment intensity as a trait that can be measured on a continuous scale and that is conditioned by many different genes and the environment.
Diversify your teaching team: most educators did not learn genetics this way, nor did their teaching assistant(s). If your instructional team has limited experience or confidence in teaching quantitative genetics, consider working in a team with genetics expertise. In this study, our instructional team included expertise in ecology and genetics. We produced instructional materials together, team-taught class meetings and lab sections, and solicited feedback from one another on how we discussed fundamental concepts with students. We particularly focused on using precise language and shared formative assessment and observations, thereby limiting possibilities for introduction of known misconceptions.
Limitations of Study
The word associations were a helpful exploratory tool and revealed intriguing shifts in concepts used by students in their explanations of phenotypic variation. Using word association in combination with the heuristic of threshold concepts offered potential probes and unique insight into students’ cognitive structures and served as a focus for analysis of student explanations. On the other hand, word associations alone could not provide insight on students’ understanding or how students’ connected the concepts in a specific way. Model-based reasoning, as detailed in Hmelo-Silver and Pfeffer (2004) and, more recently, in Dauer et al. (2013), may further our capacity to study students’ development of systems thinking and their capacity to connect concepts of variation and transition between the micromolecular to the macro scales relevant to genetics understanding.
Because we viewed this study as a first glimpse and exploration of student learning through a new piece of curriculum, we did not anticipate needing to identify and align individual student's artifacts. Our research protocol required that individual students be deidentified and that data be analyzed in aggregate form. Therefore, we could not align pre and post word associations or student explanations as we would have liked. We hope to capture this type of resolution in future studies.
CONCLUSION
Our perspective on genetics education is that students need an integrated introduction. Moreover, given the advances in genomics and proteomics, it is possible to introduce students to the broad array of tools to identify specific genetic variants as part of their research, launching them into contemporary genetics research (Redfield, 2012). Furthermore, genetics education provides an inroad to evolution that has students working with evolutionary concepts within the context of an inquiry-based curriculum—even before they are taught about evolution overtly. This notion is consistent with the American Association for the Advancement of Science Vision and Change report (2011) in support of inquiry in undergraduate biology education and, more specifically, Cunningham and Wescott (2009), who contend that in order to improve evolution education we need to move toward pedagogies that allow students to generate questions and explore data. With additional scaffolding focused on concepts of scale, randomness, uncertainty, and variation, this curriculum could offer promising new venues for studying student learning of proposed “threshold” concepts (Taylor, 2006; Ross et al., 2010) as well as other integrative learning models and provides a valuable example of theory-driven curriculum development. Moving forward, we hope to use the results of this initial study to probe further into how students connect these concepts in their reasoning and sense-making about the phenotypic variation they observe.
This study responds, in part, to Dougherty's (2009) and Redfield's (2012) challenge to integrate the genetics curriculum and provides initial evidence for student progression in learning through shifts in cognitive structure and development of explanations for why and how phenotype varies. The integrative approach allows students to apply concepts of genetics, ecology, physiology, and evolution, together with process of science skills, at different levels and at different points within a curricular sequence. We recognize the inherent value of this unit as an introduction to concepts of genetics and evolution through the context of an integrated inquiry-based laboratory unit.
FOOTNOTES
1See online bibliography compiled and maintained by Dr. Michael Thomas Flanagan at University College London: www.ee.ucl.ac.uk/∼mflanaga/thresholds.html.
2See Appendix 1 in Supplemental Material 1 for a full description of RCBr seed stock of parents and F1 populations, breeding schema, control growing conditions, and a link to online video instruction on how to prepare Bottle Biology (soda bottle system) as growing containers for planting with RCBr seed (www.youtube.com/watch?v=eEOCRz0j6iA).
3Note: seed stocks used in this curriculum are experimental and are therefore only available from the Rapid-Cycling Brassica Collection (https://rcbc.wisc.edu). Seed stock numbers are as follows: P1, 1–37; P2, 1–67; F1, 1–155; F2d, 1–157; F2s, 1–158.
ACKNOWLEDGMENTS
This work was supported by the Biocore and Wisconsin Fast Plants programs at the University of Wisconsin–Madison. The authors extend our sincere thanks to Tara Davenport, Heather Smith, and Katalin Dósa for input and assistance in implementing the curriculum; to Steve Nelms for technical data management support; to Philipp Simon, Irwin Goldman, and Rosemary Russ for their thoughtful advice; and, of course, our deep appreciation to the many Biocore students who participated in this study and continually inspire us.



