A Web Application for Generation of Random DNA Sequences with a Single Open Reading Frame: Exemplars for Genetics and Bioinformatics Education
To the Editor:
We wish to bring a new Web resource to the attention of CBE—Life Sciences Education readers.
When being introduced to the central dogma of nucleic acid transactions, students are often required to identify the 5′→3′ DNA template strand in a double-stranded DNA (dsDNA) molecule; transcribe an antiparallel, complementary 5′→3′ mRNA; and then translate the mRNA codons 5′→3′ into an amino acid polypeptide by means of the genetic code table. Although this algorithm replicates the molecular genetic process of protein synthesis, experience shows that the series of left/right, antiparallel, and/or 5′→3′ reversals is confusing to many students when worked by hand. Students may also obtain the “right” answer for the “wrong” reasons, as when the “wrong” DNA strand is transcribed in the “wrong” 3′→5′ direction, so as to produce a string of letters that “translates correctly.”
In genetics and bioinformatics education, we have found it more intuitively appealing to demonstrate and emphasize the equivalence of the mRNA to the DNA sense strand complement of the template strand. The sense strand is oriented in the same 5′→3′ direction and has a sequence identical to the mRNA, except for substitution of thymidine in the DNA for uracil in the mRNA. It is thus more computationally efficient to “read” the polypeptide sequence directly from this strand, with mental substitution of thymidine in the triplets of the genetic code table. (By definition, “codons” occur only in mRNA: the equivalent three-letter words in the DNA sense strand may be designated “triplets.”) This is the same logic used in DNA “translation” software programs.
A further constraint often imposed on dsDNA teaching exemplars is that five of the six possible reading frames are “closed” by the occurrence of one or more “stop” triplets, and only one is an open reading frame (ORF) that encodes an uninterrupted polypeptide. We designate this the “5&1” condition. The task for the student is to identify the ORF and “translate” it correctly. Other considerations include correct labeling of the sense and template DNA strands, their 5′ and 3′ ends (and of the mRNA as required), and the amino (N) and carboxyl (C) termini of the polypeptide.
Thus, instructors face the logistical challenge of creating dsDNA sequences that satisfy the “5&1” condition for homework and exam questions. Instructors must compose sequences with one or more “stops” in the three overlapping read frames of one strand, while simultaneously creating two “stopped” frames and one ORF in the other. We have explored these constraints as an algorithmic and computational challenge (Carr et al., 2014). There are no “5&1” exemplars of length L ≤ 10, and the proportion of exemplars of length L ≥ 11 is very small relative to the 4L possible sequences (e.g., 0.0023% for L = 11, 0.048% for L = 15, 0.89% for L = 25). This makes random exploration for such exemplars inefficient.
We therefore developed a two-stage recursive search algorithm that samples 4L space randomly to generate “5&1” exemplars of any specified length L from 11 ≤ L ≤ 100. The algorithm has been implemented as a Web application (“RandomORF,” available at www.ucs.mun.ca/~donald/orf/randomorf). Figure 1 shows a screen capture of the successive stages of the presentation. The application requires JavaScript on the computer used to run the Web browser.
Figure 1. Successive screen captures of the webapp RandomORF. First panel: the Length parameter is the desired number of base pairs. Second panel: Clicking the “Generate dsDNA” button shows the dsDNA sequence to be solved, with labeled 5′ and 3′ ends. The button changes to “Show ORF.” Third panel: A second click shows the six reading frames, with the ORF highlighted. Here, the ORF is in the sixth reading frame on the bottom (sense) strand. The polypeptide sequence, read right to left, is N–EITHLRL–C, where N and C are the amino and carboxyl termini, respectively. The conventional IUPAC single-letter abbreviations for amino acids are centered over the middle base of the triplet; stop triplets are indicated by asterisks (*).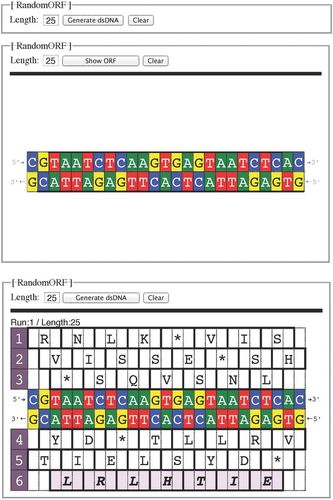
The webapp provides a means for students to practice identifying ORFs by efficiently generating many examples with unique solutions (Supplemental Material); this can take the place of the more standard offering of a small number of set examples with an answer key. The two-stage display makes it possible for problems to be worked “cold,” with the correct ORF identified only afterward. For examinations, any exemplar may be presented in any of four ways, by transposing
the top and bottom strands and/or reversing the direction of the strands left to right. Presentation of the 5′ end of the sense strand at the lower left or upper or lower right tests student recognition that sense strands are always read in the 5′→3′ direction, irrespective of the “natural” left-to-right and/or top-then-bottom order. We intend to modify the webapp to include other features of pedagogical value, including constraints on [G+C] composition and the type, number, and distribution of stop triplets. We welcome suggestions from readers.
ACKNOWLEDGMENTS
S.M.C. and H.T.W. were supported by NSERC Discovery Grants during development of the algorithm and webapp. S.M.C. thanks his students in Biol4241 Advanced Genetics, Winter 2014, for feedback on the use of the webapp.



