Beyond the Central Dogma: Model-Based Learning of How Genes Determine Phenotypes
Abstract
In an introductory biology course, we implemented a learner-centered, model-based pedagogy that frequently engaged students in building conceptual models to explain how genes determine phenotypes. Model-building tasks were incorporated within case studies and aimed at eliciting students’ understanding of 1) the origin of variation in a population and 2) how genes/alleles determine phenotypes. Guided by theory on hierarchical development of systems-thinking skills, we scaffolded instruction and assessment so that students would first focus on articulating isolated relationships between pairs of molecular genetics structures and then integrate these relationships into an explanatory network. We analyzed models students generated on two exams to assess whether students’ learning of molecular genetics progressed along the theoretical hierarchical sequence of systems-thinking skills acquisition. With repeated practice, peer discussion, and instructor feedback over the course of the semester, students’ models became more accurate, better contextualized, and more meaningful. At the end of the semester, however, more than 25% of students still struggled to describe phenotype as an output of protein function. We therefore recommend that 1) practices like modeling, which require connecting genes to phenotypes; and 2) well-developed case studies highlighting proteins and their functions, take center stage in molecular genetics instruction.
INTRODUCTION
Students enter college introductory biology courses knowing that genes determine traits, a general concept largely emphasized in primary and secondary school science (Lewis et al., 2000; Marbach-Ad, 2001; Thörne and Gericke, 2014). The causal relationship between genes and traits, however, often represents a “black box.” Few students can provide mechanistic explanations of how gene expression leads to protein production, let alone of how protein function may lead to a phenotype (Marbach-Ad and Stavy, 2000; Lewis and Kattmann, 2004). In addition, students struggle to understand how genetic variation arises and how it results in phenotypic variation (Bray Speth et al., 2014).
Genetics, more specifically molecular genetics, is reportedly challenging for learners in high school and college (Bahar et al., 1999; Lewis et al., 2000; Marbach-Ad and Stavy, 2000; Marbach-Ad, 2001; Lewis and Kattmann, 2004; Duncan and Reiser, 2007); the emerging consensus in the literature is that much of this difficulty can be explained by the large amount of technical vocabulary genetics requires and by its multilevel nature. Multilevel thinking, that is, the ability to integrate concepts across different levels of biological organization, is intrinsically difficult for learners, as it requires understanding how subcellular and cellular (microscopic) mechanisms and interactions bring about observable (macroscopic) traits (Marbach-Ad and Stavy, 2000). An alternative model of multilevel thinking in genetics is proposed by Duncan and Reiser, who posit that genes exist on distinct ontological levels: genes are simultaneously physical entities (nucleotide sequences occupying specific locations within chromosomes) and units of genetic information that code for proteins. To make sense of how genes function, students must conceptually integrate genes’ informational and physical properties (Duncan and Reiser, 2007). Heredity adds yet another layer of complexity to the gene concept, as learners can think about genes from a within-generation perspective, that is, genes influencing the phenotypes of the organisms carrying them; and from a between-generations perspective, that is, genes influencing the phenotypes of their progeny. Students, however, often focus on only one of these at a time (Marbach-Ad, 2001; Tsui and Treagust, 2010).
The difficulties with molecular genetics reflect some of the challenges that learners encounter when dealing with systems. Systems have been broadly defined as entities that are composed of multiple structural components and that exist and function as wholes through the interactions of their parts (Ben-Zvi Assaraf and Orion, 2005; Jacobson and Wilensky, 2006). Systems typically encompass multiple hierarchical levels of organization and exhibit feedback loops, nonlinear dynamics, and emergent properties. Learning and reasoning about systems present significant cognitive challenges, because they often require connecting phenomena across multiple levels of organization and explaining macrolevel properties or outcomes through microlevel, hidden causal mechanisms (Hmelo-Silver and Azevedo, 2006; Jacobson and Wilensky, 2006; Liu and Hmelo-Silver, 2009). In the context of molecular genetics, we can view cells, organisms, and populations as systems in which genetic information is stored, expressed, and transmitted. Organisms’ phenotypes, for example, emerge as a result of processes and networks of interactions occurring at the molecular, cellular, tissue, and organ levels, and are often influenced by environmental factors (Marbach-Ad and Stavy, 2000; Marbach-Ad, 2001; Duncan, 2007; van Mil et al., 2013). Furthermore, phenotypic variation (a property of populations) has its origins in genetic mutation (a molecular-level causal mechanism).
While there is no single framework to define what abilities are representative of systems thinking, multiple frameworks, developed in different educational contexts, converge on several key skills (Boersma et al., 2011). For instance, systems thinking was characterized by Verhoeff et al. (2008), in the context of cell biology education, as a suite of abilities to: 1) distinguish among biological levels of organization and match concepts to their specific level; 2) interrelate concepts within the same level of organization (horizontal connections) and across levels of organization (vertical connections); and 3) apply general, abstract system models to specific concrete instances, and vice versa (Verhoeff et al., 2008; Boersma et al., 2011). In the context of elementary earth science education, systems thinking was characterized as neither innate nor one-dimensional but as developing over time with practice, in a hierarchical progression of increasingly sophisticated skills that build upon one another (Ben-Zvi Assaraf and Orion, 2005). These hierarchical skills were adapted to describe learning about biological systems, specifically the human body, and grouped into three sequential levels, each one forming the basis for the following (Ben-Zvi Assaraf et al., 2013):
Level A: Analysis—the ability to identify structural components and processes within a system | |||||
Level B: Synthesis—includes the ability to identify simple (structural) and dynamic (functional) relationships among the system’s components, and to organize components and processes into a network of relationships | |||||
Level C: Implementation—the ability to recognize “hidden” system components and causes, make generalizations, and think temporally about a system, both retrospectively and predictively | |||||
It has become evident in recent years that traditional science education is not conducive to acquisition and development of systems-thinking skills, because instruction is primarily focused on fact accretion with little effort made to promote interconnectedness of concepts (Jacobson and Wilensky, 2006). In line with current awareness of these shortcomings and of the need for a more integrative approach, the latest recommendations for biology education incorporate a focus on systems and cross-cutting competencies like modeling, at both the K–12 and the college levels (American Association for the Advancement of Science [AAAS], 2011; Next Generation State Standards [NGSS] Lead States, 2013). As attention to systems and systems thinking in the science classroom increases, so does the need for adequate instructional approaches and for suitable methods of assessment (Boersma et al., 2011; Brandstädter et al., 2012). Concept maps have been investigated as possible conceptual representations that reveal some elements of systems thinking (Sommer and Lücken, 2010; Brandstädter et al., 2012). However, concept maps are a tool for representing declarative knowledge, that is, what the author knows about a domain or system; they are, thus, static representations that fail to convey higher-order systems-thinking skills, such as thinking temporally and abstractly (Vattam et al., 2011; Tripto et al., 2013). Sommer and Lücken (2010) used concept maps as a way of eliciting students’ system organization abilities, but other forms of assessment (e.g., open-response questions) were needed to elicit system properties, such as emergence and cause–effect relationships. A more appropriate means of prompting systems thinking is to have students represent systems as physical, computational, or conceptual models (Verhoeff et al., 2008; Evagorou et al., 2009; Liu and Hmelo-Silver, 2009; Honwad et al., 2010). Model-based learning (Gobert and Buckley, 2000) is an instructional approach that optimally addresses the cognitive challenges posed by systems. Models, as abstract or conceptual representations of systems, allow learners to focus on the relevant structures and mechanisms operating within a system (Jordan et al., 2008; Verhoeff et al., 2008; Liu and Hmelo-Silver, 2009; Long et al., 2014). Model building engages learners in an “intentional, dynamic and constructive process” (Jonassen et al., 2005, p. 15) that mediates meaningful learning, intended as the gradual construction and reorganization of one’s mental knowledge structures. We adopted a conceptual system-modeling strategy rooted in structure–behavior–function (SBF) theory, which was originally developed to describe complex engineered systems (Goel et al., 1996). According to SBF theory, all systems are composed of a multitude of individual components (the structures of the system) that interact with one another in a network of interrelationships (the behaviors of the system) to produce an output (the function of a system). SBF theory has been adapted to create conceptual modeling practices suitable for teaching and learning about biological systems at the middle school and college levels (Hmelo-Silver and Pfeffer, 2004; Jordan et al., 2008; Liu and Hmelo-Silver, 2009; Goel et al., 2010; Vattam et al., 2011; Long et al., 2014). Student-generated SBF models of biological systems have been used to investigate college introductory biology students’ reasoning about the origin of variation and evolution (Dauer et al., 2013; Bray Speth et al., 2014).
With this study, we sought to use systems thinking as a framework to develop instruction and assessment and as a lens to interpret evidence of student learning of how genes determine phenotypes. In a large-enrollment introductory biology course for majors, we implemented a learner-centered, model-based instructional design. Guided by theory and evidence on hierarchical development of systems-thinking skills, we scaffolded instruction so that students would focus first on defining the relationships between pairs of molecular genetics structures (i.e., between DNA and RNA, between gene and protein, etc.), a task that maps onto the most basic systems-thinking skill set, analysis (Table 1). Next, students started integrating these structures and relationships to build generalized conceptual models of gene expression in the cell, which required advancing to the synthesis stage of systems thinking. Modeling prompts became gradually more complex over time to include additional structures, functions, and levels of organization, toward the final goal of developing gene-to-phenotype models that conveyed: 1) the origin of genetic variation in a given population and 2) how phenotypes within that population are determined by the organisms’ genotypes. While the analysis and synthesis levels of systems thinking could be easily mapped onto learning outcomes, such as articulating relationships among pairs of biological structures (analysis) and integrating all these relationships into a functional network (synthesis), the highest level (implementation) remained more elusive. We approached implementation by asking students to apply their general model of gene expression to multiple real-world scenarios. By articulating models contextualized to the function of specific genes with defined phenotypes, students practiced generalizing their understanding of the gene-to-phenotype paradigm to explain an array of instances in which information contained in genes is expressed and results in phenotypes.
| Systems-thinking ability | Model-based instruction practice |
|---|---|
| Analysis: ability to identify relationships among the system’s componentsa,b | Scaffolding: students articulate the relationships between pairs of structures. |
| Synthesis: ability to organize the systems’ components and processes into a framework of relationshipsa,b | Model building: students are provided a list of structures and are required to 1) connect them in a meaningful network, 2) articulate the relationships among them, and 3) select and represent processes that are relevant to the function of the system. |
| Implementation: ability to make generalizations;a,b ability to move back and forth between general models of systems and concrete biological systemsc | Contextualizing: students apply the general variation-to-phenotype framework to model how different, specific systems work. |
We adapted the systems-thinking hierarchy as a lens for analyzing students’ artifacts and as a framework for organizing our research questions. In an introductory biology course implementing model-based and case-based instruction on the central dogma of molecular biology, do students improve over time in their ability to 1) correctly articulate individual relationships between relevant biological structures; 2) organize these relationships into a meaningful model that explains how genes determine phenotypes; and 3) modify their language, when required, to make their models context-specific?
We hypothesized that students would rapidly acquire the basic ability to identify and articulate relationships among structures that mediate expression of genetically determined phenotypes (DNA, gene, allele, mRNA, protein). We expected that integrating structures and relationships into meaningful networks and contextualizing language to make models case specific, would develop gradually and become more refined over time, as they require more advanced systems-thinking skills.
METHODS
Course Context and Pedagogy
This research was conducted in one semester of a large-enrollment majors’ introductory biology course at a large private research university in the midwestern United States. The course introduced students to principles of cell and molecular biology, genetics, and animal form and function. Students (n = 129) were primarily freshmen (84%). Biology majors/minors represented ∼44% of the population, and 43% of all students had taken AP Biology in high school. Sixty-five percent of all students reported they intended to pursue premedical studies. The class met each week for two 75-min periods for 15 wk.
A single instructor (E.B.S.) taught the course and implemented an active, learner-centered pedagogy that frequently engaged students in constructing conceptual system models, as described in previous studies (Dauer et al., 2013; Bray Speth et al., 2014). Instruction in this course was a pilot application of the “flipped-classroom” model (Bull et al., 2012; Herreid and Schiller, 2013), informed by the SBF theory of systems and by evidence on the cognitive affordances of systems thinking. Students were expected to learn information about biological structures before coming to class, by studying instructor-recorded screencasts and assigned readings and by completing homework quizzes and modeling activities. In class, students worked in permanent collaborative groups of three. Class time was devoted to elucidating biological mechanisms and processes through small-group discussion of preclass assignments, worksheets and clicker problems, instructor explanations, and whole-class discussions.
This research was reviewed and determined exempt by the local IRB (protocol 16795). All course students received a recruitment statement early in the semester and had the opportunity to opt out of the study if they wished their data not to be included in analyses for research purposes.
Timeline of Instruction and Assessment
Introduction to Modeling (Weeks 1 and 2).
Students learned early in the semester how to represent biological systems as semantic box-and-arrow networks, colloquially referred to in class as SMRF (structure–mechanism/relationship–function) models. Modeling was introduced in week 1 through a classroom activity that involved reading and discussing a paragraph about nitrification in aquaria (Supplemental Materials S1). Aquaria as examples of complex biological systems were previously used to learn and implement SBF modeling in educational settings (Hmelo-Silver and Pfeffer, 2004; Vattam et al., 2011). In our course, students were asked to identify, in the text provided, the structures composing the aquarium system and to represent them on paper as nouns in boxes. Next, they were asked to connect the boxes with labeled arrows to represent how, through interactions among the structures, the system accomplished the nitrification function. Students were encouraged to talk through these tasks with their neighbors; however, everyone turned in an individual model on carbonless paper to receive completion credit. With students’ input, the instructor built a consensus model on the board. At the beginning of the following class period, in week 2, the instructor reviewed the activity and shared with students a set of practical SMRF model-building conventions (Supplemental Materials S2), which served as reference for all subsequent modeling activities throughout the semester.
Molecular Genetics Pretest (Week 3).
In weeks 2 and 3, students learned about principles of chemistry, structure and function of biological molecules, cell membranes and cell structure. At the end of week 3 (before instruction on the central dogma of molecular biology), students completed a pretest (in the last 10–15 min of class time) intended to gauge their existing knowledge and understanding of key molecular genetics structures (DNA, mRNA, protein, chromosome, gene, and allele) and of the relationships between them. One of the tasks on the pretest (adapted from Marbach-Ad, 2001) was as follows:
Complete the sentences below with verbs or phrases that communicate your best understanding of the relationships between these pairs of biological structures:
DNA _____ mRNA
mRNA _____ Protein
Gene _____ DNA
Gene _____ Protein
Allele _____ Gene
Hereafter, we will refer to these as the fill-in-the-blank PW (PW) relationships. This task was subsequently incorporated verbatim on exam 1 (in week 5) and again on the cumulative final exam (week 16).
Gene-to-Phenotype Instruction and Modeling (Weeks 4 and 5).
Instruction focused on gene expression (transcription and translation), mutation, and formation of new alleles. One of the key learning outcomes was that students would be able to construct a model that explains both how genes mutate and how they are expressed to determine phenotypes.
We collected the most common student answers to the fill-in-the-blank pretest task and used them to build an online homework assignment formulated as a set of “check all that apply” items, such as the following:
DNA ______ mRNA.
a. transcribes
b. is transcribed by
c. is transcribed into
d. codes for
e. converts into
f. contains
Because the course-management system we used at the time did not allow a clear distribution of partial credit, we initially chose to only mark as wrong the obviously incorrect choices (like “e” and “f,” above). The homework was thoroughly reviewed in class, providing the opportunity to explicitly discuss what appeared to students as lexical nuances (e.g., why is “transcribed into” a better choice than “transcribed by”?), thus differentiating between correct and marginal choices. No answer key was provided in the postclass slides, so students had to study from the notes they took in class; the fill-in-the-blanks task, however, was re-presented to students on exam 1 and, at that point, direct feedback was provided to each student on whether each of his or her statements was accurate, inaccurate, or marginal (only accurate answers received full credit).
Students created their first generalized DNA-to-protein model in week 4 as homework, in response to the following prompt: “Build a box-and-arrow (SMRF) model representing your current understanding of how genetic information contained in DNA is used to make proteins. At a minimum, use the following five structures: DNA, nucleotides, mRNA, protein, amino acids.” At the beginning of the following class, students turned in one copy of their preclass homework models (to earn completion credit) and kept one copy for in-class discussion with their collaborative teams and note taking. The instructor guided groups’ conversations by reminding students of the key conventions of model construction (Supplemental Materials S2), and answered any questions emerging from the discussion. After this activity, the instructor proceeded to explain to the whole class the structure of genes and the basic mechanics of transcription and translation. A follow-up homework assignment (Supplemental Materials S3) required students to revise their models based on their new knowledge and understanding, to articulate in writing what they changed in their models and why, and to add the new structure, “gene,” to their revised models.
In week 5, as a collaborative group activity in class, students extended their models to include the concepts “allele” and “phenotype.” This activity was contextualized within the case study of the mouse Mc1R gene, which encodes a plasma membrane receptor critical for determining fur color (www.lbc.msu.edu/evo%2Ded/; White et al., 2013); this case allowed explicit connection of gene/allele expression to phenotype at the organismal scale. The activity prompt was: “Build a model that illustrates how the light brown coat phenotype originated in mice. Use: DNA, RNA, gene, allele, protein, phenotype. Make the language specific to the case we analyzed.” Feedback was provided to the whole class at the end of the activity. At the end of the activity, the instructor guided a whole-class discussion by having groups report out on 1) how they contextualized the wording in their models, 2) how they incorporated allele, and 3) how they included phenotype in their models. The instructor summarized the consensus generated by the students and annotated the class slides, which were provided to students after class. The instructor-generated summary was a short list of tips: 1) an example of contextualization (e.g., “protein” in the general model becomes “Mc1r protein” in the contextualized model, 2) new alleles are a direct result of genetic mutation, and 3) phenotype expression is an output of protein function. These notes did not include examples of whole models, leaving students free to develop their own without preconceptions of what a model should look like.
Exam 1 (Week 5).
Exam 1, administered at the end of week 5, incorporated a case study about toxin resistance in clams (www.lbc.msu.edu/evo%2Ded/; White et al., 2013; Supplemental Materials S4) and included multiple-choice application questions, a contextualized gene-to-phenotype model, and the fill-in-the-blanks task (five PW relationships, same as on the pretest). The prompt for the model-building task was as follows:
Illustrate how genotype determines phenotype in toxin-resistant clams. Use, at a minimum, the following structures: DNA, gene, allele, mRNA, protein, phenotype. Make sure you incorporate the mutation event.
After grading the exam but before returning it to students, the instructor implemented a classroom activity aimed at discussing the exam gene-to-phenotype model and providing general feedback. Students had to recreate the model in their collaborative groups (one per group). One group was called to report out; they drew their model on the whiteboard and the whole class had a discussion, specifically focused on whether the model 1) appropriately incorporated the mechanism of mutation and 2) was contextualized to the clam case.
Final Exam (Week 16).
The final cumulative exam, in week 16, incorporated a case study about obesity in mice (Supplemental Materials S4). Questions based on this case study included a set of multiple-choice application items and a gene-to-phenotype model-building task. The model prompt was as follows:
Illustrate (a) how the obese phenotype originated in the wild-type mouse population, and (b) how genotype determines phenotype in the ob (obese) mice. Use, at a minimum, the following structures: DNA, gene, allele, mRNA, protein, phenotype.
In addition, students were again asked to complete the same five fill-in-the blank PW relationships as on exam 1.
A summary of the course timeline and relevant assessments can be found in Figure 1. A timeline of all molecular genetics SBF model-building tasks throughout the course of the semester can be found in the Supplemental Material (Supplemental Table S1). Multiple-choice questions developed for the case-based exams will be made available to readers upon request.
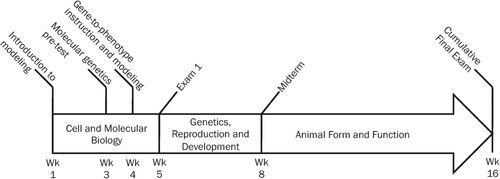
Figure 1. Timeline of course instruction and relevant assessments.
Data
Fill-in-the-Blank PW Relationships.
We collected students’ responses to the fill-in-the-blank task, which was presented on the pretest (week 3) and incorporated verbatim on exam 1 (in week 5, immediately following instruction on the central dogma of molecular genetics) and on the cumulative final exam (week 16). All student responses were transcribed into Microsoft Excel for analysis.
Models.
We analyzed gene-to-phenotype models generated by students on exam 1 and on the final exam. The model prompts for the two exams differed in context (Supplemental Table S1). The expected model functions, however, were equivalent, and the list of provided structures was identical. Gene-to-phenotype model prompts were preceded in both exams by 1) text that explicitly described for each case the wild-type phenotype and the mutation leading to a new phenotype and 2) a set of multiple-choice questions that probed students’ reasoning about biological concepts in the context of the cases. Background text and model prompts for both exams are included as Supplemental Materials S4.
Data Analysis
Propositional Accuracy Rubric.
Students were taught that each “box-arrow-box” group (i.e., each “proposition”) within their models should be a complete, coherent statement that should be meaningful on its own. Throughout this study, we refer to propositions as the smallest meaningful units in which a student model can be decomposed; similarly, for the fill-in-the-blank task, each structure-blank-structure is a proposition.
We developed a single rubric to rate biological accuracy of students’ propositions, applicable to both the decontextualized fill-in-the-blank relationships and the propositions within models.
To generate and validate a coding rubric grounded in students’ answers, we identified a set of frequent responses to fill-in-the-blank relationships generated by students on the pretest, and we added some frequent relationships extracted from exam 1 gene-to-phenotype models. Ten individuals (six biology faculty, two graduate students, and two undergraduate students who were members of the biology education research team) independently assigned scores to all propositions, using the following scale, similar to that used in Dauer et al. (2013):
0 = missing | |||||
1 = incorrect, inappropriate (e.g., “DNA turns into mRNA”) | |||||
2 = marginally correct, ambiguous, or poorly worded (e.g., “DNA is used to produce mRNA”) | |||||
3 = as accurate as can be expected from an introductory biology student after instruction (e.g., “DNA is transcribed into mRNA”) | |||||
To generate a consensus rubric, we averaged the scores assigned by the 10 independent raters to each proposition and determined ranges as follows: propositions that received a mean score <1.5 were categorized as 1 (beginning); propositions that received a mean score between 1.5 and 2.5 were categorized as 2 (developing); propositions that received a mean score >2.5 were categorized as 3 (mastered). The rubric for propositional accuracy of relationships is available as Supplemental Table S2.
Because not all possible student answers were included in the set coded by 10 individuals, we performed an interrater reliability procedure, to account for responses requiring the rater to make a judgment call. Two raters independently applied the rubric to code greater than 30% of the fill-in-the-blanks PW relationships from the pretest. The interrater agreement was high for all five PW relationships (Cohen’s kappa: 0.883–1.000). Because of the high degree of agreement, one rater coded the rest of the PW relationships.
When rating propositions within students’ models, because students could draw the arrows in any direction, we adapted the rubric to all “reverse relationships” (i.e., we coded an mRNA → DNA relationship based on the rubric for the DNA → mRNA relationship). Two raters independently coded greater than 30% of the PW relationships within the exam 1 gene-to-phenotype model. The interrater agreement was high for all PW relationships (Cohen’s kappa: 0.817–0.947). A single rater coded the rest of the PW relationships within students’ models.
Model Explanatory Power Rubric.
We identified four key processes—mutation, transcription, translation, and phenotype expression—as essential to convey a complete and coherent account of the origin of variation and flow of genetic information (i.e., the functions of the gene-to-phenotype model). We developed a rubric (Table 2) to code models based on whether they included these four processes and whether the processes were appropriately connected within the model to represent the flow of information from genetic variation to phenotype (i.e., mutation causes change in the nucleotide sequence; that change in information is passed from gene to mRNA via transcription, resulting in translation of a different protein; protein function leads to phenotype expression).
| Processb | Criteria for “presence” | Criteria for “appropriate connection” |
|---|---|---|
| Mutation | The model includes the word “mutation” or otherwise describes a change in the information, e.g., “DNA, if copied incorrectly, will cause a change in the gene.” | The mutation, or change, is shown as directly affecting the gene/allele or nucleotide sequence/DNA. |
| Transcription | The model includes the word “transcription” or one of its derivatives or an otherwise acceptable synonymous expression, e.g., “DNA serves as a template for mRNA.” | The model clearly indicates that information is transferred from DNA/gene/allele to mRNA. |
| Translation | The student uses the word “translation” or one of its derivatives or an otherwise acceptable synonymous expression, e.g., “mRNA codes for a protein.” | The model clearly indicates that information is transferred from mRNA to protein. |
| Phenotype expression | The model incorporates the word “phenotype” or its case-specific description. | Phenotype is represented as an outcome of protein function (as opposed to a direct output of gene or allele, for example). |
Two raters independently applied the rubric to code 36 student models not included in this data set. The mean percent agreement between the two raters was 95.1% for all four processes (range: 93.1–97.2%). Because of the high level of agreement, one rater coded all the models for this analysis.
Language Consistency and Contextualization.
We analyzed and compared students’ language when articulating individual PW relationships in the fill-in-the-blank format and within gene-to-phenotype models, on both exam 1 and the final exam. We considered language to be consistent if the student used exactly the same words on both the fill-in-the-blank and model on the same exam or used the passive form of the same words (e.g., a student stated that “a gene is made up of DNA” on the fill-in-the-blank assessment and then wrote that “DNA makes up a gene” in the model, on the same exam).
Model prompts required students to modify their language, to make it specific to each case. We analyzed how frequently students contextualized propositions within models. We coded a proposition as contextualized if the behavior or at least one structure was modified to be case specific, for example, “mouse allele,” “Mc1R protein,” “toxin-resistant phenotype,” or “transcribed into mRNA with ob mutation.” Each model could contain multiple instances of contextualization, at distinct levels of biological organization. We found that students may add specificity to their language when representing structures/relationships at the molecular level (i.e., by naming the specific gene or allele), at the cellular level (i.e., by naming the protein), and/or at the organismal level (i.e., by specifying the phenotype or at least by naming the organism). Two raters independently coded 40 gene-to-phenotype models from exam 1 for contextualization at the three levels of organization. Interrater agreement was 95% for the molecular level and 87.5% for the cellular and organismal levels. The two coders discussed discrepancies and came to a consensus to refine further coding. The same procedure was applied to final exam models: two raters independently coded 40 gene-to-phenotype final exam models. Interrater agreement was 95% for the molecular level, 100% for the cellular level, and 97.5% for the organismal level. Owing to the high level of agreement, a single rater coded the remaining models for both exams.
Statistical Analyses
We analyzed change in accuracy of students’ propositions (both the decontextualized PW fill-in-the-blanks and the propositions within models) across assessments at two or three time points (see Results). Propositional accuracy was measured on an ordinal 0–3 scale, and class data were not normally distributed; therefore, we applied repeated-measures, nonparametric statistical tests. A Friedman test followed by post hoc Wilcoxon signed-rank tests was used to compare propositional accuracy of fill-in-the-blank items across three time points (pretest, exam 1, and final exam); the Wilcoxon signed-rank test was applied directly to compare accuracy of propositions within models on exam 1 and the final exam.
We also compared, across exam 1 and the final exam, the proportion of students who incorporated and correctly connected propositions within models and the proportion of students who contextualized the language of their models. To determine whether these proportions changed from exam 1 to the final exam, we applied McNemar’s test, which is used to determine whether there is a difference in the distribution of a dichotomous variable in a repeated-measures design. All statistical analyses were performed in IBM SPSS Statistics, version 21.
RESULTS
Students’ Ability to Accurately Articulate Relationships between Pairs of Molecular Genetics Structures Develops Early and Persists Throughout the Semester
We compared the accuracy of students’ language in the fill-the-blank PW relationships across three time points: pretest (before molecular genetics instruction, week 3), exam 1 (after molecular genetics instruction, week 5), and final exam (end-of-the-semester cumulative final, week 16). Students who completed all three assessments were included in this analysis (n = 115). We applied a Friedman test for each fill-in-the-blank PW relationship to determine whether individual students performed similarly at each of the three time points (repeated-measures design). All Friedman tests (Table 3) identified significant differences among students’ accuracy scores at the three time points. Post hoc Wilcoxon signed-rank tests with Bonferroni correction (α = 0.025) showed that, for each of the five relationships, accuracy of students’ statements increased significantly from pretest to exam 1 but did not significantly change from exam 1 to the final exam (Table 3).
| Mean propositional accuracy ± SD | Friedman test (α = 0.05) | Post hoc Wilcoxon signed-rank tests (α = 0.025) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Relationship | A. Pretest | B. Exam 1 | C. Final exam | χ2 (df = 2) | p | Z (A–B) | p(A–B) | Z(B–C) | p(B–C) |
| DNA_mRNA | 1.21 ± 0.83 | 2.35 ± 0.65 | 2.37 ± 0.71 | 136.9 | <0.001 | −8.226 | <0.025 | −0.486 | 0.627 |
| mRNA_protein | 1.24 ± 0.96 | 2.43 ± 0.69 | 2.37 ± 0.78 | 106.5 | <0.001 | −7.741 | <0.025 | −0.892 | 0.373 |
| Gene_DNA | 1.77 ± 0.99 | 2.27 ± 0.64 | 2.27 ± 0.71 | 29.0 | <0.001 | −4.488 | <0.025 | −0.064 | 0.949 |
| Gene_protein | 0.93 ± 0.97 | 2.30 ± 0.85 | 2.43 ± 0.87 | 131.2 | <0.001 | −7.981 | <0.025 | −1.613 | 0.107 |
| Allele_gene | 1.09 ± 0.94 | 2.11 ± 0.96 | 2.27 ± 0.98 | 95.9 | <0.001 | −6.962 | <0.025 | −1.857 | 0.063 |
To investigate whether the plateau in accuracy from exam 1 to the final exam was due to students memorizing and repeating the same connecting words, we compared individual students’ responses to all five PW fill-in-the-blank questions across the two exams (exam 1 and the final). Surprisingly, the percentage of students using the same wording on both exams was relatively low and varied among the five PW relationships (ranging from 25.2% for gene_DNA to 41.7% for DNA_mRNA).
Students’ Models Incorporate Individual Relationships with Varying Frequency and Accuracy
We analyzed gene-to-phenotype models created by students on exam 1 and the final exam to find out: 1) how frequently students incorporated, in their models, the propositions they articulated in a fill-in-the-blank format on the same exam; 2) whether students used the same wording across decontextualized, stand-alone relationships and models; and 3) whether the accuracy of propositions within models was the same as that of the fill-in-the-blank PW propositions.
Students incorporated all five given PW relationships in their models, with a wide range of frequencies (Table 4). The gene_protein relationship was the least frequently incorporated on both exams. Interestingly, the frequency of all other relationships within gene-to-phenotype models changed from exam 1 to the final exam, with some relationships decreasing in frequency (e.g., DNA_mRNA) and others increasing (e.g., mRNA_protein). Moreover, we observed that, when incorporating any given relationship into their models, most students did not use the same wording they used to complete the corresponding fill-in-the-blank relationship (Table 4).
| Exam 1 model | Final exam model | ||||
|---|---|---|---|---|---|
| Relationship | Frequency | Same wording | Frequency | Same wording | |
| DNA_mRNA | 70.4% | 40.9% | 53.0% | 31.3% | |
| mRNA_protein | 67.0% | 48.3% | 78.3% | 43.5% | |
| Gene_DNA | 85.2% | 40.9% | 73.0% | 19.1% | |
| Gene_protein | 13.9% | 7.8% | 13.9% | 7.8% | |
| Allele_gene | 78.3% | 34.7% | 74.8% | 22.6% | |
Accuracy of students’ language in the fill-in-the-blank PW relationships on exam 1 was significantly better than that of the same relationships within gene-to-phenotype models for all except the DNA_mRNA and gene_protein propositions (Wilcoxon signed-rank test, p < 0.05, two-tailed). On the final exam, accuracy of students’ language was again significantly lower in the model than in the fill-in-the-blank PW relationships for the gene_DNA and allele_gene propositions but not for the other three propositions (Table 5).
| Exam 1 accuracy | Final exam accuracy | |||||
|---|---|---|---|---|---|---|
| Relationship | n | Fill-in-the-blank | Model | n | Fill-in-the-blank | Model |
| DNA_mRNA | 81 | 2.35 | 2.20 | 61 | 2.28 | 2.28 |
| mRNA_protein | 77 | 2.40 | 2.19* | 90 | 2.47 | 2.39 |
| Gene_DNA | 98 | 2.29 | 1.75* | 84 | 2.37 | 1.87* |
| Gene_protein | 16 | 2.63 | 2.19 | 16 | 2.19 | 2.00 |
| Allele_gene | 90 | 2.19 | 1.76* | 86 | 2.31 | 1.91* |
Students’ Models Reflect Difficulties Representing and Appropriately Incorporating the Origin of Variation and the Protein-to-Phenotype Connection
We analyzed gene-to-phenotype models to determine whether students incorporated and appropriately connected the processes of mutation, transcription, translation, and phenotype expression (see Table 2 for the rubric).
Students consistently incorporated transcription and translation in their models with high frequency and appropriately placed these processes within the context of genetic information flow. The origin of variation and phenotypic expression appeared more problematic; although students incorporated the processes with relatively high frequencies, they often failed to appropriately connect them within their models on both exam 1 and the final exam (Table 6).
| Exam 1 | Final exam | |||
|---|---|---|---|---|
| Incorporated (%) | Appropriately connected (%) | Incorporated (%) | Appropriately connected (%) | |
| Mutation | 79.2 | 52.2 | 87.8 | 74.8* |
| Transcription | 93.9 | 90.4 | 95.7 | 95.7 |
| Translation | 88.7 | 87.0 | 87.8 | 87.8 |
| Phenotype Expression | 87.8 | 53.0 | 96.5 | 71.3* |
The frequency with which students appropriately connected the processes of mutation and phenotype expression in their gene-to-phenotype models increased significantly from exam 1 to the final exam (Table 6; McNemar’s test, p < 0.05, two-tailed).
Students Struggled to Contextualize Their Gene-to-Phenotype Models
We investigated to what extent students contextualized their models by analyzing whether they modified structures or behaviors to make them context specific. We organized our analysis of model contextualization using levels of biological organization. On exam 1, almost half the students contextualized the phenotype (organismal level). Fewer students contextualized the gene/allele (molecular level) and protein (cellular level), and only 25.2% of students mentioned the organism’s name (Table 7). We compared the frequency at which students contextualized the language of their models on exam 1 and the final exam. For all levels of biological organization, including mention of the organism’s name, we found a significant increase in the proportion of students who contextualized their language (Table 7; McNemar’s test, p < 0.05, two-tailed). Despite this significant improvement, almost one-third (32.2%) did not contextualize gene/allele on either exam, and 46.1% did not contextualize the protein on either exam.
| Organism | Molecular level (gene) | Cellular level (protein) | Organismal level (phenotype) | ||
|---|---|---|---|---|---|
| Exam 1 | Context | Clam | Na+ channel gene/allele | Na+ channel protein | Resistance to toxin |
| Frequency | 25.2% | 23.5% | 22.7% | 47.8% | |
| Final exam | Context | Mouse | ob gene/allele | Leptin | Obesity |
| Frequency | 41.7% | 60.0% | 47.0% | 78.3% |
DISCUSSION
In this study, we investigated students’ learning about how genes determine phenotypes over the course of one semester of introductory biology. Design of model-based instruction and assessment was informed by our current understanding of how learners develop systems-thinking skills. Specifically, we aimed to create learning activities that were aligned with the hierarchical sequence of systems-thinking abilities as described in the literature (Table 1). Mirroring the cognitive progression from analysis to synthesis and implementation, we first engaged students in articulating relationships between pairs of molecular genetics structures (DNA, gene, mRNA, protein, and allele). Next, students practiced organizing these structures and relationships into networks of interactions by constructing conceptual models illustrating how genetic information codes for proteins. Over time, modeling prompts gradually grew in complexity, ultimately requiring students to illustrate the origin of genetic variation and the expression of a new phenotype in a population. Students constructed multiple gene-to-phenotype models, contextualized to various case studies. While learners in this study rapidly acquired the ability to correctly articulate individual, isolated relationships between molecular genetics structures, model building presented an additional challenge, as it required students to integrate their understanding of multiple biological structures and processes into a single explanatory framework. Learning to articulate accurate, meaningful, context-specific gene-to-phenotype models required time and repeated bouts of practice and feedback.
Model-Based Practices Elicit and Reveal Systems-thinking Abilities
The most basic systems-thinking skill set is analysis: the ability to identify components (structures and processes) within a system and the relationships among them (Ben-Zvi Assaraf and Orion, 2005; Ben-Zvi Assaraf et al., 2013). In this introductory-level biology course, we expected students to articulate in their own words the relationships between structures. A pretest, administered before instruction, provided information on students’ baseline ability to convey five core molecular genetics relationships. Students’ ability to accurately articulate the relationships between pairs of molecular genetics structures significantly improved early on and persisted throughout the semester (Table 3). Students completed the same five relationships on exam 1 and the final exam; comparison between these two exams revealed that most students used different wording to articulate the same relationships. This suggests that most students did not memorize the relationships but continued to fine-tune their vocabulary over time.
A more advanced systems-thinking skill set, resting upon knowledge of structures and processes, is synthesis: the ability to organize a system’s components within a meaningful framework of relationships. In the context of our course, model building was the practice associated with this skill set. By scaffolding (atomizing the gene-to-phenotype process into a suite of PW relationships; Table 1), we had intended to provide a stepping-stone for students to help them with model building. We did not, however, suggest to students that they could or should have used these relationships to “seed” their models. Articulating the network of relationships that explains a function of the system appears to be a more difficult task than filling in the blanks (as we would expect), based on the overall lower accuracy of relationships within models, compared with the individual PW relationships (Table 5). Another way of looking at this is that, just as a system is more than the sum of its parts, a system model is more than the sum of a given set of relationships. Propositions, as “fragments,” may be easy to formulate in isolation but less easy to integrate into a meaningful whole. This may explain why the accuracy of students’ propositions within models was in some cases lower than the accuracy of the same relationships articulated as stand-alone fill-in-the blank statements. Our focus on model explanatory power, further discussed later, represents an initial attempt at capturing a student model as a whole, by analyzing the ability of the model to convey the required functions with appropriate choice and organization of the key processes.
Another interesting finding was that, on both exam 1 and the final, students did not include in their models all possible PW relationships but made choices (Table 4). For example, the direct gene-to-protein relationship was incorporated within models very infrequently, we think because students represented the central dogma of molecular genetics (DNA → mRNA → protein) in two steps, making it unnecessary or redundant to also directly connect gene to protein. It is also important to note that we only report the frequency with which students included in their models the five relationships for which we also have fill-in-the-blank data. Student models, however, included a whole variety of other relationships they chose to articulate, like gene to mRNA or allele to mRNA, which might explain the drop in the frequency of incorporation of the DNA-to-mRNA relationship in their models from exam 1 to the final exam (Table 4). Overall, our analysis of students’ gene-to-phenotype models indicates that these types of artifacts are flexible and allow a great deal of choice, being in a sense idiosyncratic.
The third and most advanced set of systems-thinking skills, implementation, refers to learners’ ability to “use” models to describe concrete instances, to make inferences, to reason retrospectively and to make predictions. While implementation may be achieved in multiple ways and clearly represents a suite of skills, in this study we only focused on students’ ability to move from general to specific, represented by construction of multiple case-based gene-to-phenotype models. Students started by developing generalized models of gene expression, then were asked to create context-specific models grounded within case studies to explain how variation for a given trait came about in a population. The ability to move from generalized models to concrete instances and vice versa was described in other studies on learning about systems as a higher-order systems-thinking ability (Verhoeff et al., 2008; Ben-Zvi Assaraf et al., 2013). In different educational settings and when learning about different natural systems, instruction and learning may begin with general patterns and move toward specific cases, or vice versa. Either way, we argue that generalization and contextualization are two sides of the same coin. The goal of this practice is to give learners multiple opportunities to apply and transfer the same general principle to multiple instances, which is considered to be a way of anchoring science knowledge into concrete examples, thus promoting more robust understanding (Hmelo and Narayanan, 1995). In our case, we found that our students’ ability to construct context-specific gene-to-phenotype models was limited at first but improved significantly over the course of the semester (Table 7). Further research will be necessary to more clearly articulate the various facets of the implementation level of systems thinking, specifically what activities and products constitute, respectively, appropriate practice and evidence of this type of thinking.
Students’ Gene-to-Phenotype Models Improve with Practice and Feedback
In this course, students received individual feedback only on the PW relationships and models they created on exams; the instructor graded exams and shared the rubric with the class, to enable students to interpret their scores. Students, however, had repeated opportunities throughout the semester to evaluate models they built, both individually and in their collaborative groups. For example, a homework activity assigned early in the semester (review–reflect–revise) required students to review their preclass gene-to-phenotype models, reflect on what they had learned in class, revise their models, and explain in writing what they changed in their models and why. In class, students constructed context-specific gene-to-phenotype models in their collaborative groups. After model construction, the instructor prompted students to discuss whether their models conveyed the requested functions, and to report out for class discussion. Emphasis on model function was an explicit target of instruction and feedback. Students were repeatedly asked “Does your model tell a story?” or “Does your model show the origin of variation and phenotype expression in this system?” Models/representations of knowledge that emphasize function over structure are indicative of expertise development; novices build more fragmented (chunked) models but fail to meaningfully link chunks together to show how the system produces its outputs (Hmelo-Silver and Pfeffer, 2004; Moss et al., 2006). Moss et al. (2006) measured the “functionality” of engineering students’ representations of artificial systems, where functionality was intended as models’ coherence and ability to convey the requested functions. These authors related model functionality and internal connectivity to students’ early stages of expertise development: seniors’ models were more “functional” and coherent than those of freshmen (Moss et al., 2006). Schwarz et al. (2009) used a similar measure when investigating middle school students’ representations of natural systems, but they called it “explanatory power.” We wanted to analyze the explanatory power of our students’ models as a measure of how well they convey the system’s function, thus moving the focus beyond analysis of the accuracy of individual relationships. We observed that, while students’ gene-to-phenotype models initially conveyed an understanding of the centrality of transcription and translation (central dogma of molecular genetics), the origin of genetic variation was represented less frequently, and both the origin of variation and phenotype expression were often inappropriately integrated within the model (Table 5 and Figure 2A). Student models’ explanatory power significantly improved from exam 1 to the final exam, in which a larger proportion of student models incorporated these mechanisms in the appropriate logical order (Table 6 and Figure 2B), suggesting that students, over time, became better able to convey with their models the required functions of origin of genetic variation and phenotype expression, articulating relationships that occur beyond the core mechanisms of the central dogma. We had previously observed in a different student population that students struggled with modeling the genetic origin of variation, in the context of evolution teaching and learning (Bray Speth et al., 2014). Aware of this difficulty, we designed the prompt for the model on exam 1 to include an explicit requirement to illustrate the origin of variation (“Make sure you incorporate the mutation event”). This scaffold was removed from the prompt on the final exam (see Methods); still, the frequency with which students incorporated and appropriately connected mutation as the source of genetic variation significantly increased on the final exam (Table 6).
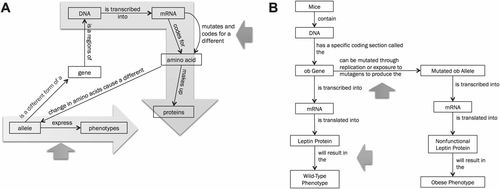
Figure 2. Examples of student-generated gene-to-phenotype models from exam 1 (A) and the final exam (B). Models were transcribed verbatim from the original, handwritten student work, in black and white. We added colored arrows to point out specific model features relevant to our analysis. Orange arrows represent where students incorporated mutation, and green arrows point at phenotype expression. Two light blue–shaded arrows superimposed on model A highlight how students would often develop two distinct but incomplete branches in their models, one from gene/allele to protein, the other from gene/allele to phenotype.
In this study, we evidence difficulties with representing phenotype expression as a direct outcome of protein function. Several factors may contribute to these difficulties. First, in traditional secondary science education, learning about gene expression and learning about traits are often compartmentalized and occur at different times; certainly, this compartmentalization is evident in textbooks (Flodin, 2009). Classical genetics focuses on genes as determinants of hereditary traits, whereas molecular biology focuses on genes coding for proteins (Lewis and Kattmann, 2004; van Mil et al., 2013). Oftentimes, the bridge between these two areas of biology, specifically the role of proteins as gene products that influence or determine phenotypic traits, is not made explicit, leaving students with a branched, fragmented view. Studies of “teachers’ talk” about genes in high school pointed out how, due to the compartmentalization of school curricula, the resulting gene conceptions may be fragmented into discrete, disconnected gene-to-protein and gene-to-traits schema (Marbach-Ad, 2001; Thörne and Gericke, 2014). Although exam 1 occurred before instruction on classical genetics, most of our students had high school biology, so we assume that they might have been exposed to genetics in high school. This may explain the patterns we observed in students’ gene-to-phenotype models on exam 1. The students who failed to connect protein to phenotype often developed branched models: one branch represented the gene (or allele) coding for the protein, while the other branch represented a direct gene/allele → phenotype connection (Figure 2A). These results are consistent with those reported by Marbach-Ad (2001), who observed a similar branching pattern when 12th-grade students were asked to build concept maps connecting various genetics structures.
Proteins and Their Functions Need to Take the Stage in Molecular Genetics Instruction
Duncan (2007) proposed that reasoning about how genes determine phenotypes is based on a few key heuristics: genes code for proteins, proteins as central, and effects through interaction (the latter meaning that molecular genetics processes require physical interactions between components). Our instruction focused on the first of these (genes code for proteins), which highlights how information contained in genes is used in the cell to produce specific proteins. The assumption learners might derive is that, if a gene influences a phenotype, then this phenotype must be caused or mediated by a protein. This is, of course, a major limitation and, as we know, only offers a partial explanation for most phenotypes in nature. While we are fully aware of further layers of complexity brought by multigenic traits, regulatory DNA regions, epigenetic control, and non protein-coding genes (to name a few), these variations on the theme of genetic control of phenotype are most typically the subjects of upper-division courses. Although some more complex cases could definitely be accessible to introductory-level students, in our course we strove to keep the focus on basic examples in which a known mutation in a single protein-coding gene leads to a specific phenotype. Our data clearly show that students struggled to articulate the direct connection between protein and phenotype (Table 6). This observation is consistent with a previous study showing that high school students failed to incorporate the concept of protein when explaining genetic phenomena (Duncan and Reiser, 2007). Part of this difficulty can be explained by lack of knowledge and understanding of what proteins “do” in the cell and how their activities explain emergent phenotypes (Duncan, 2007; Duncan and Reiser, 2007). From a systems-thinking perspective, defining a phenotype in terms of protein function requires learners to leap from a subcellular-level structure to an emergent property at the organismal level. If learners have not first practiced describing the outcomes of protein function at the cellular level, such a direct leap to phenotype may appear daunting and inexplicable. This difficulty may contribute to an explanation of the direct link between genes/alleles and phenotype we observe in many students’ models (Figure 2A).
Implications for Teaching and Learning
Sustainability.
One of our course goals was that students would routinely engage in higher-order learning activities, including building conceptual gene-to-phenotype models, both in and out of class. This approach obviously leads to a large amount of student-generated artifacts and poses the challenge of providing adequate and constructive feedback, especially since modeling tasks are also presented in high-stakes exams. A well-recognized challenge for instructors of large courses is the limited ability to provide individualized feedback on complex constructed-response tasks. Our approach was to design activities that leveraged students’ self-evaluation and peer-evaluation skills: in-class and homework model-building activities were regularly followed by self-evaluation prompts (review–reflect–revise) and by guided small-group and whole-class discussion. Students’ accountability was achieved by giving credit for all in-class and homework activities, including reflections and model revisions. The instructor only provided individualized, direct feedback to every student for models produced on exams (high-stakes assessments). This approach allowed implementing model-based instruction in a large class by a single instructor without a grader or graduate teaching assistant; the grading load was sustainable, since the responsibility of evaluating low-stakes assignments was distributed between the teacher and the learners. The data collected and presented in this study, unfortunately, do not allow parsing out the relative contributions of class-level and individual-level feedback to explain improvement of learners’ modeling abilities over time. We aim at addressing this question in future studies specifically designed to analyze student response to different modes of practice, instructor feedback, and self-evaluation. Potential avenues for prompting students’ reflection on their own model may include, among others, an analysis of whether the language used in their models reflects the accuracy of the language they used to formulate individual PW relationships.
Focus on Information Flow, Systems, and Modeling.
In this study, we used a model-based approach to infuse a systems-thinking perspective into teaching and learning about a core concept of biology, information flow (i.e., the origin of genetic variation and how genes determine phenotypes). While “information flow, exchange, and storage” has traditionally represented a staple of introductory biology courses, recent calls like Vision and Change challenge instructors to adopt active and integrative approaches and to incorporate concepts like systems and cross-cutting competencies like modeling, at the K–12 and the college levels (AAAS, 2011; NGSS Lead States, 2013). Our approach to teaching and learning about information flow in first-semester introductory biology is informed by these recommendations and integrates learning about the flow of genetic information with systems and modeling. Systems thinking, with its emphasis on illuminating how emergent properties at higher levels of organization result from dynamic interactions at lower levels, perfectly applies to the gene-to-phenotype relationship.
Case Studies Are Integral to Learning How Genes Determine Phenotypes.
Case studies that require explaining phenotypes in terms of molecular and cellular processes should be the centerpiece, not an afterthought, of molecular genetics instruction. Examples should, in fact, “drive the learning” (Duncan, 2007), as they lend themselves to constructing meaningful and authentic accounts of reasoning, like explanations and models, and they allow learners to apply their knowledge, which otherwise remains abstract, disconnected, and superficial (Spiro et al., 1988). In light of the literature, our course experience, and the results of this study, we advocate incorporating case studies as instructional activities that not only help students learn what functions proteins perform but, most importantly, that facilitate acquisition of the proteins-as-central heuristic, whereby students can assume the key role of a protein when proposing a plausible molecular explanation for a phenotype (Duncan, 2007). Several studies have already advocated for a strategic use of case studies to generate a deep and meaningful understanding of the connection between molecular biology, genetics, and evolution (Kalinowski et al., 2010; White et al., 2013). A case-based approach to biology should permeate the curriculum to provide students multiple examples and opportunities to make the types of connections that most textbooks fail to make explicit (Spiro et al., 1992) but that are necessary for an integrated understanding of biology.
ACKNOWLEDGMENTS
We are very thankful to many friends and colleagues. Brian Downes, Jon Fisher, Blythe Janowiak, Laurie Russell, Laurie Shornick, Ranya Taqieddin, Kolin Clark, and Zisansha Zahirsha contributed to validation of the propositional accuracy rubric. Ranya Taqieddin also tested the rubrics used in this study, participating to establish interrater reliability. We thank Joe Dauer, Tammy Long, Jenni Momsen, Laurie Russell, and members of the Bray Speth lab for critically reviewing the manuscript and Lauren Arend for her advice on statistical analysis. This article is based on work supported in part by the National Science Foundation (NSF awards 0910278 and 1245410). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NSF.



