Development of the Central Dogma Concept Inventory (CDCI) Assessment Tool
Abstract
Scientific teaching requires scientifically constructed, field-tested instruments to accurately evaluate student thinking and gauge teacher effectiveness. We have developed a 23-question, multiple select–format assessment of student understanding of the essential concepts of the central dogma of molecular biology that is appropriate for all levels of undergraduate biology. Questions for the Central Dogma Concept Inventory (CDCI) tool were developed and iteratively revised based on student language and review by experts. The ability of the CDCI to discriminate between levels of understanding of the central dogma is supported by field testing (N = 54), and large-scale beta testing (N = 1733). Performance on the assessment increased with experience in biology; scores covered a broad range and showed no ceiling effect, even with senior biology majors, and pre/posttesting of a single class focused on the central dogma showed significant improvement. The multiple-select format reduces the chances of correct answers by random guessing, allows students at different levels to exhibit the extent of their knowledge, and provides deeper insight into the complexity of student thinking on each theme. To date, the CDCI is the first tool dedicated to measuring student thinking about the central dogma of molecular biology, and version 5 is ready to use.
INTRODUCTION
Well-designed assessment tools are essential for instructors to evaluate class-level understanding of a particular topic before instruction, to measure the effectiveness of their lessons, and to test new tools and methods in the classroom. In essence, good assessment tools allow transformed classrooms to evolve based on evidence. In the 20+ yr since the introduction of the Force Concept Inventory (Hestenes et al., 1992), dozens of other concept assessment tools have been created for many science, technology, engineering, and mathematics (STEM) fields, including physics (e.g., Thornton, 1998; Singh and Rosengrant, 2003; Ding et al., 2006), statistics (Stone et al., 2003), geosciences (Libarkin and Anderson, 2005), and engineering (e.g., Midkiff et al., 2001; Krause et al., 2003; Steif and Dantzler, 2005). The number of concept assessments on biological topics is quickly expanding, as more instructors and researchers recognize the need for research-based tools to evaluate student understanding of essential biological concepts (e.g., Kalas et al., 2013; Abraham et al., 2014; Deane et al., 2014; Price et al., 2014; Williams and Heinrichsen, 2014; Couch et al., 2015a). Many of the more recently developed instruments align with some of the five core concepts required for biological literacy as described in the National Science Foundation/American Association for the Advancement of Science 2009 report Vision and Change: A Call to Action (AAAS; 2011).
One of these core ideas, “Information Flow, Exchange and Storage,” not only pertains to many topics covered in college biology classrooms but is arguably the basis for all modern genetic and genomic research. The concept of information being permanently stored in DNA, transiently copied into RNA intermediates, and used to build proteins that carry out the majority of cellular functions, is recognized as the “central dogma of molecular biology” (Crick, 1970). The topic is visited many times over the course of a biology curriculum. The biology education research literature supports the notion that typical undergraduate biology students struggle with concepts related to information flow (Pashley, 1985; Stewart et al., 1990; Allchin, 2000; Lewis and Wood-Robinson, 2000; Lewis et al., 2000; Marbach-Ad, 2001; Khodor et al., 2004; Wright and Newman, 2013; Wright et al., 2014), perhaps due to high school curricula failing to prepare students for deep learning on these important topics (Lewis et al., 2000; Shaw et al., 2008).
In teaching courses such as introductory biology, cell biology, molecular biology, and genetics, instructors often struggle to identify and fill in the apparent gaps in student knowledge and help make clear connections between key topics that students often miss when trying to learn biology, especially in the context of information flow. While instruments such as the Genetics Concept Assessment (Smith et al., 2008), the Genetics Literacy Assessment (Bowling et al., 2008), the Biology Concept Inventory (Klymkowsky et al., 2010), the Meiosis Concept Inventory (Kalas et al., 2013), and the Molecular Biology Capstone Assessment (Couch et al., 2015a) each include some questions that relate to information flow, to date there is no dedicated assessment instrument focused on the central dogma. Informal discussions with colleagues and a review of available assessment tools confirmed our belief that this is a tool much needed by the community.
Good assessment tools do not just differentiate students with “correct” versus “incorrect” ideas; they can also be used to identify expert-like thinking about a particular topic. Developmental and cognitive psychologists describe disciplinary experts as those with deep content knowledge who are also able to adapt, organize, and apply knowledge in a dynamic and meaningful way (Newell and Simon, 1972; Bédard and Chi, 1992; Chi, 2006). One of the overarching goals of undergraduate science education is to promote expert-like thinking and reasoning skills in students as they progress through curricula and college programs. Unfortunately, research has shown that many assessments used in higher education classrooms test little more than factual recall and/or require that students only use low-level cognitive skills to come up with correct answers (Palmer and Devitt, 2007; Crowe et al., 2008; Momsen et al., 2010). Think-aloud interviews and individual oral examinations often show underlying conceptions and thought processes that are not apparent with a typical multiple-choice test or even open-ended written questions (Dufresne et al., 2002; Chi, 2006; Kuechler and Simkin, 2010), but most instructors do not have the time to evaluate each student so deeply. Thus, carefully constructed and tested instruments like the Central Dogma Concept Inventory (CDCI) are needed to more accurately identify student ideas and evaluate development of disciplinary expertise in the typical college student.
A major obstacle to learning biology, and especially genetics, is the large amount of vocabulary and its precise usage for communicating concepts clearly (Pearson and Hughes, 1988; Groves, 1995; Bahar et al., 1999). Novices, however, may be able to recognize or produce correct terminology or phrases without necessarily understanding the deep concepts linked with the terms. For example, we have interviewed students who state, “DNA is a template,” or “DNA is copied,” without being able to correctly define the term “template” or explain any of the specific molecular interactions or processes that facilitate replication. Novices also may interchange or incorrectly substitute terminology, even though their underlying knowledge about the concept is correct. For example, in our research, a biology student once described the first step of gene expression as “Translation is when single strand DNA is copied to form RNA strands.” Although the student correctly articulated that a new molecule of RNA was synthesized during the process, s/he mixed up the terms “transcription” and “translation.” It would be interesting to know how such an answer would be graded by instructors—do they typically put more emphasis on correctness of vocabulary or correctness of concept?
Perhaps the most difficult type of response to evaluate comes from students who use vague language when asking and/or answering questions in class or on assessments. Not only does imprecise student (novice) language make instructors cringe, but it makes interpreting the statements or written responses difficult. In many cases, student-generated answers are not quite correct, but not totally incorrect either, which puts the burden of interpretation solely on the grader. Vague language often results in an instructor 1) wondering whether a particular student actually knows the material but just cannot articulate it and/or 2) giving students the “benefit of the doubt” and rewarding them for superficial and/or incorrect ideas. Research has demonstrated that vague and imprecise student language is an issue in many STEM settings (Kaplan et al., 2009; Peterson and Leatham, 2009; Haudek et al., 2012; Rector et al., 2012; Wright et al., 2014). Students often misapply words that have nuanced meanings to experts and/or use nonscientific definitions of terms when describing scientific phenomena. When corrected or confronted by an instructor, students often say, “That’s what I meant,” probably not realizing how shaky their foundational knowledge actually is.
In this paper, we describe the development and testing of the CDCI, which extends and builds on a long-term research project focused on student understanding of genetic information flow. We specifically focus on the vague and imprecise language that students use when describing aspects of information flow and turned it into items in our assessment tool, following a similar framework to what has been described by others in the discipline-based education research community (e.g., Garvin-Doxas and Klymkowsky, 2008; Adams and Wieman, 2011; Abraham et al., 2014; Deane et al., 2014; Price et al., 2014; Towns, 2014). To gain insight into how undergraduate students think about the essential topics, we used a variety of sources, including classroom artifacts, written responses to open-ended questions, and interviews with students at all undergraduate levels (Wright and Newman, 2013; Wright et al., 2014). Thus, we were able to identify a wide range of conceptions (correct as well as incorrect) and language (accurate as well as inaccurate) that students use to describe the central dogma. We used the strategy of incorporating students’ own language and incorrect ideas as distractors to ensure that the assessment captures the true ideas that students hold about biological phenomena and not just what educators think their students know (Garvin-Doxas and Klymkowsky, 2008; Klymkowsky and Garvin-Doxas, 2008).
Concept inventories are usually constructed as a series of multiple-choice items designed to probe students’ conceptual knowledge about a particular topic. While we initially constructed the CDCI as a forced-choice assessment, early in development we changed the format to a multiple-select instrument. A multiple-select format circumvents many of the issues associated with a forced-choice format that can result in students using “test-taking strategies” to eliminate incorrect choices and help them guess the correct answer without advanced content knowledge (Haladyna and Downing, 1989; Haladyna et al., 2002; Towns, 2014). In addition, we found it difficult to construct biologically relevant questions within the constraints of having one absolutely correct answer and three to four absolutely incorrect answers, especially without including artificial distractors, whereas the new format broadened the range of questions we could ask. Finally, this format allowed for multiple levels of understanding to be incorporated in a single question, which makes it applicable to individuals with a wide range of sophistication and reveals both breadth and depth of their knowledge (Couch et al., 2015a). In this paper, we present our strategies and methodologies for the construction of the CDCI tool. We also discuss specific examples of how student ideas and language were incorporated into the instrument and present examples of question evolution based on validation interviews and expert feedback. Results from a large-scale beta test (n = 1733) are presented and discussed. We also present evidence to demonstrate instrument reliability (Cronbach’s alpha > 0.8, which indicates good internal consistency) and support the hypothesis that the CDCI measures student understanding of central dogma concepts (the overall score on the CDCI increases when test takers have more biology course experience, and a class focused on central dogma–related topics showed improvement in postcourse testing).
METHODS
All student data were collected with proper institutional review board approval. Student participants for versions 1–3 were biology majors from a large, private, STEM-focused institution in the northeast United States. Participants for testing of version 4 were from nine different institutions, discussed in more detail below.
Overview of Instrument-Development Process
Owing to the iterative nature of developing assessment tools, the CDCI has gone through several major revisions (Table 1). Version 4 (CDCI v.4) underwent large-scale beta testing with more than 1700 students, to allow for a final revision to CDCI v.5, which is now available for general use. While the CDCI was originally designed as a standard multiple-select tool, perspectives and opinions from focus-group discussions led to a reevaluation of all items. Items were rewritten to accommodate a multiple-select instrument in CDCI v.2 that was maintained in all later versions.
| CDCI version | Format of item responses | Description |
|---|---|---|
| 1 | Twenty-four items | Tool was constructed after analysis of student data (class artifacts, open-ended responses, interviews). |
| Multiple choice | Items and responses were rewritten to accommodate a multiple-select format after informal focus-group discussion with research team and collaborators. | |
| 2 | Twenty-four items | Version 2 was used in think-aloud interviews with 12 undergraduate (sophomore, junior, and senior) biology students. Field notes and interview transcripts promoted revisions to the CDCI. |
| Multiple select | ||
| 3 | Twenty-four items | Version 3 was taken by 54 undergraduate (sophomore, junior, and senior) biology students recruited from a number of different courses (molecular biology, biotechnology, and bioinformatics courses). |
| Multiple select | Version 3 was reviewed by 13 biology experts (PhD faculty at a number of institutions) using an online survey format. Experts answered CDCI questions and provided feedback on question clarity and relevance. | |
| Analysis of expert feedback, deeper analysis of student interview transcripts, and CDCI scores prompted revisions to the CDCI. | ||
| 4 | Twenty-four items | Large-scale beta-test version administered at eight institutions to a total of 1733 students (for more information, see Table 2). |
| Multiple select | ||
| 5 | Twenty-three items | Ready for classroom use. Contact authors if interested in using the CDCI v.5. |
| 5 | Multiple select |
The conceptual framework for development of the CDCI is shown in Figure 1. Initially, a large list of student-derived ideas from classroom observations, class artifacts, and research data were grouped into themes. Themes were used to identify six major concepts related to the central dogma (Table 2). The concepts were used as a basis for construction of preliminary items, in which distractors and language were guided by student ideas. Several CDCI items were developed from each major concept. Student feedback, in the form of pilot testing and think-aloud interviews, was used to improve items in versions 1–3 (CDCI v.1–3). Expert feedback, in the form of surveys, supported our choices of the major concepts and helped improve items in CDCI v.3 to create CDCI v.4. Large-scale testing and statistical analysis was used to revise version 4 into its current, ready-to-use version (CDCI v.5).
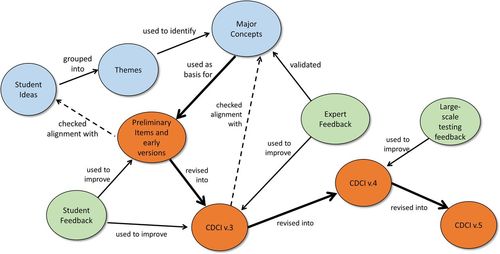
Figure 1. Flowchart of CDCI item-development process. Versions of the instrument are shown in orange, sources of content are blue, and sources of feedback are green. Following the thick arrows shows the evolution of the instrument; thin arrows show how components were used to support the CDCI development; and dotted lines indicate checks.
| Major concept | Corresponding CDCI questions | Major concept | Corresponding CDCI questions |
|---|---|---|---|
| Macromolecules are composed of specific building blocks. | 3. Which of the following chemical groups are identical in DNA and RNA? | There are multiple types of information encoded in DNA that may be used at different times. | 6. For a typical eukaryotic gene, which of the following is normally expressed as part of the protein? |
| 17. Are amino acids and nucleotides considered to be macromolecules or building blocks? [building blocks vs. macromolecules] | 5. Which of the following does a typical human gene contain? | ||
| 7. Which of the following statements correctly describes a promoter? [encoded in DNA, not RNA or attached later] | |||
| 21. When are noncoding regions removed from a gene? | |||
| Mechanism of RNA synthesis. | 1. Which of the following molecules is needed in order for a cell to carry out transcription? | Mechanism of protein synthesis. | 2. Which of the following molecules is needed in order for a cell to carry out translation? |
| 10. How is a region of double-stranded DNA molecule changed during the process of transcription? | 4. In which of the following processes does a nucleic acid exhibit catalytic activity? | ||
| 16. Which of the following describes the process of transcription? [template and building blocks] | 14. Which of the following must occur during the process of translation? [molecular interactions] | ||
| 13. What do mRNA, tRNA and rRNA have in common? | 16. What role does mRNA play in the process of protein synthesis? | ||
| 20. During the protein synthesis, amino acids are bound to which molecules? | |||
| Although mistakes can occur in any central dogma process, mutations are permanent changes in the DNA. | 19. Imagine that you identify a mutation in a human cell line. Which of the following must also be true? [mutations do not always affect products] | DNA is permanent information storage and products (RNA and proteins) are synthesized when needed. | 8. You are comparing brain, heart, liver, and skin cells from the same individual. Which of the following molecules would you expect to be identical between these four cell types? |
| 22. An error in which of the following processes could result in a heritable mutation? [only replication] | 9. A cell receives a signal that it needs to produce a certain protein for the first time. Which of the following processes must occur? | ||
| 23. An error in which process can result in a non-functional protein product? | 11. Which of the following functions does double-stranded DNA play in a bacterial cell? | ||
| 12. Which of the following functions does mRNA play in a bacterial cell? | |||
| 18. What do replication and transcription have in common? |
Our design strategy adhered to the principles of construct validity. According to Messick (1989, 1995), the major concerns about validity are construct underrepresentation (the assessment is too narrow to accurately measure understanding) and construct-irrelevant variance (the assessment is too broad in that extraneous ideas can be used to answer questions). These two overarching ideas informed the design of the CDCI. For example, the use of the multiple-select format can address both concerns: individual questions are not overly narrow, because different levels of expertise or facets of knowledge can be represented in a single item, and they are not overly broad, because test takers have to consider every response independently rather than using extraneous cues to eliminate possibilities. The iterative process of evaluating each item allowed us to ensure that interpretation of items fell into the intended range.
Details of the Process
Because the primary developers of the CDCI had been involved in several projects investigating student understanding of central dogma–related topics (Wright and Newman, 2013; Wright et al., 2014) and had taught courses in introductory biology, cell biology, molecular biology and genetics, both course artifacts and research data were used to construct the CDCI. To select and define the concepts to be included in our concept inventory, we created a large, preliminary list of incorrect or unclear statements made by students when discussing the central dogma. For example, students repeatedly exhibited difficulty in differentiating macromolecules (nucleic acids and proteins) from their building blocks (nucleotides and amino acids). They also talked about promoters as if they were RNA or protein rather than being part of the DNA, or were added onto a gene rather than being incorporated into the genome from the beginning. A partial list of ideas that we determined were problematic for biology students includes:
Building blocks of macromolecules (DNA, RNA, protein) are not interchangeable.
Amino acids are covalently linked together during protein synthesis.
Genes are part of genomes, whether or not they are expressed.
End products of DNA replication, transcription, and translation are new macromolecules.
Translation cannot occur unless the process of transcription has already occurred.
Different types of RNA molecules (mRNA, tRNA, rRNA) have specific roles.
Promoters are sequences of DNA.
tRNA molecules are the functional products of tRNA genes.
This list was eventually condensed and edited to a list of six major concepts related to the central dogma that seem to be the most problematic for students (Table 2).
To create CDCI items, we examined written responses to open-ended questions and interview transcripts from research projects investigating student understanding of information flow to try and uncover the underlying thought processes that gave rise to their words. We then created items with concrete options (correct answers and distractors) based on the possible right and wrong ideas that might lead students to use vague statements. We believe this method to be effective, because accuracy in interpretation of student language is evaluated later in the testing process. If we misinterpreted student ideas and created unrealistic distractors, then students would not choose those options during beta testing, and the response would be discarded as a result. Table 3 shows an example of how student language was interpreted and used to develop a question and response choices.
 |
Thirteen external faculty reviewers (all of whom had a PhD in biology or a related field) were asked for feedback on whether any important concepts were missed in CDCI v.3, but none were identified. One reviewer pointed out that the tool did not include gene regulation and that this would be a good subject for a future instrument. We agree; the CDCI is limited to the basics of gene expression and intentionally does not address the more advanced topics of gene regulation. The final CDCI questions were later realigned with each of the main concepts to ensure that each concept was linked with at least two questions. Originally, there were eight main concepts, but after review of the alignments by six additional external faculty experts and further discussion among the research team, they were reworded and condensed to the final six presented here (Table 2).
Validation Interviews with CDCI Questions (Version 2)
Think-aloud interviews, using CDCI v.2, were conducted with 12 biology majors (sophomore, junior, and senior levels). Students were asked to read each question and response choice aloud and talk their way through how they would answer each question. Interviews were video-recorded, and interviewers took field notes to help them remember interesting things students said and to flag questions that seemed especially problematic or confusing to students. Video files were later uploaded into NVivo 10 (QSR International), so interviews could be transcribed and synced with video files. The NVivo software package allowed the research team to code interesting student comments and identify potentially problematic wording. These observations were used to revise the instrument to CDCI v.3.
Pilot Testing (Version 3) with Students and Biology Experts
CDCI v.3 was pilot tested with 54 students majoring in biology or a related subject such as biotechnology or biomedical sciences (13 sophomores, 22 juniors, and 19 seniors). Student participants were recruited from several different laboratory sections of 200- to 400-level courses. Students who agreed to participate took the CDCI during “downtimes” in their laboratory sessions and typically took 15–20 min to answer all questions; no one required more than 30 min. The CDCI v.3 was scored by both partial credit (92 total responses) and by whole question (24 total responses).
To gain expert feedback, we gave an online version of the CDCI v.3 to biology faculty members of diverse institutions across the country to 1) answer the questions for comparison with developers’ intended correct answers and 2) provide feedback and suggestions on each item. The faculty members were asked to select the correct response(s) for each question and provide comments on the clarity of each question. Nine reviewers answered all questions and provided written feedback, while four reviewers did not attempt to answer any of the CDCI questions themselves but did provide feedback on question clarity and relevance. Numerical scores from CDCI v.3, reviewer feedback, and deeper analysis of interviews were used to revise the instrument to CDCI v.4.
Large-Scale Beta Testing (Version 4)
CDCI v.4 was given to 1733 students in nine classes at eight institutions, representing a diverse population in terms of geography, institutional type, and class size (see Table 4). At each test site, the CDCI was administered during class using paper and pencil. A large number of students took the CDCI as a preinstruction assessment in their introductory biology course (n = 1396), a group that we designated as “preintroductory.” A different set of students took the CDCI as a post–introductory biology assessment (n = 235), a group that we designated as “postintroductory.” A third group of students (n = 102) took the CDCI as a postcourse assessment in an intermediate-level course (genetics and cell/molecular biology) and are designated as the “postintermediate” group. One large introductory biology class accounted for 68% of the data (preintroductory group only). Individuals who gave invalid responses (e.g., left a question blank or selected choice “E” when only “A” to “D” were possible) were excluded from the reported analyses (final n = 1541).
 |
Included in the postintermediate group were the scores from 53 students in a molecular biology course who had also taken the test as a pretest (those scores are not included in the above data). To demonstrate content validity of the instrument, we also compared pre- and posttest scores of this group in a separate analysis. Because the assessment was given anonymously, it was not possible to correlate scores from individuals, and five subjects who took the pretest did not take the posttest. Therefore, we did the t test analysis two ways: first, we compared the means of the 53 posttests with those of the 58 pretests, and second, we dropped the 5 lowest pretest scores. The latter assumes that the lowest scores came from all the people who did not take the posttest and therefore gives a very conservative estimate of the difference between the two groups.
Statistical Analyses
Because preinstruction introductory biology students struggle with concepts related to genetic information flow, we assumed these very novice students had a relatively high guess rate when answering CDCI questions. To perform item analysis, we used all postinstruction CDCI scores for final statistical analyses (n = 318 valid responses). Student answers on individual items were compared with their overall scores (Ding et al., 2006; Kalas et al., 2013). Difficulty index (P) was calculated as the percentage of students who answered correctly on a given question. Discrimination (D) was calculated as the difference between the percent of students who got the question right in the top 27% of overall scores and the percent of students who got the question right in the bottom 27% of students. Point biserial correlations (rpb) indicate the extent to which score on the individual question correlates with overall score on the whole assessment. Cronbach’s alpha was calculated using the statistical package psych (Revelle, 2015) to determine internal consistency of the instrument.
RESULTS
Pilot Testing with Students and Biology Experts
After the CDCI had undergone revisions based on informal feedback and validation interviews we used the instrument (version 3) in a small-scale beta test with two distinct populations of subjects; undergraduate biology students (sophomore through senior, n = 54) and biology experts (faculty, n = 13). Owing to the nature of the instrument (multiple select), scores were calculated in two ways: 1) the percentage of 24 questions that were answered completely correctly (% whole questions correct), and 2) the percentage of total choices that were correctly marked/not marked out of 92 possible responses (% response options correct).
As shown in Figure 2, A and B, the mean scores for experts were higher than for students, with seniors scoring above underclassmen and below faculty. All data from students and faculty were combined in a single analysis of variance (ANOVA), and the trend of increasing scores with increasing level of expertise was found to be significant: whole question F(3, 59) = 13.2, p < 0.0001; response option F(3, 59) = 10.4, p < 0.0001. When groups were compared by t test, sophomores and juniors were not significantly different from each other t(33) = 0.66, p > 0.1, but all other pairs tested were significantly different (e.g., whole question scores comparing juniors with seniors t(39) = 2.78, p = 0.0084, seniors to faculty t(26) = 2.97, p = 0.0064) with a large effect size (Cohen’s d > 0.8). The lack of difference between sophomores and juniors is not surprising given the inconsistent nature of the biology curriculum at the institution where the beta tests took place. Owing to the fact that sophomores and juniors were part of the same courses targeted for beta testing, we feel that the labels “sophomore” and “junior” are simply reflective of their time in college not necessarily the amount of exposure to central dogma–related topics.
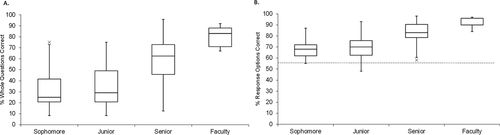
Figure 2. Results of pilot testing (CDCI v.3). The instrument was given to both undergraduate students (n = 54; 13 sophomores, 22 juniors, 19 seniors) and faculty (n = 9). (A) Each whole question, scored as fully correct or incorrect (24 questions with three to five response options each). (B) Each response option, scored independently as correct or incorrect (92 T/F options). The dotted line indicates the hypothetical guess rate (50%).
The faculty reviewers not only answered the CDCI v.3 questions but also offered comments about the accuracy and relevancy of each item. Items that were answered incorrectly by more than one expert reviewer were closely examined for grammatical or factual errors. In all cases, questions that were answered differently by multiple faculty had also been flagged by our reviewers as potentially problematic. Table 5 shows several examples of how items changed based on expert feedback; most of the changes involved rewording the questions and/or responses. Using the scores and expert comments, we further revised the CDCI into version 4, which was used in large-scale beta testing.
 |
Large-Scale Beta Testing
Faculty were recruited for beta testing of CDCI v.4 in diverse classroom settings. Students were given the test in a paper format during class time, either pre- or postinstruction on the central dogma, in introductory biology or intermediate-level courses. Based on the timing and specific class, beta testers were grouped as “preintroductory,” “postintroductory,” or “postintermediate” (Table 4). Figure 3, A and B, demonstrates that, as students gained more classroom experience, median CDCI scores increased (ANOVA test, F(2, 1538) = 37.1, p < 0.0001; t test on each pair, p < 0.0001) with a moderate to large effect size for each pair (Cohen’s d for preintroductory to postintroductory: 0.78 for whole question and 0.67 for response options; preintroductory to postintermediate: 0.74 for whole question and 0.90 for response options).
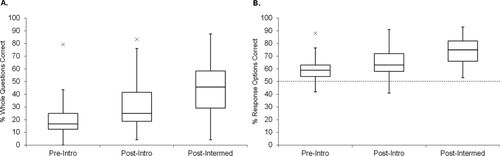
Figure 3. Results of large-scale beta testing (CDCI v.4). The CDCI was given as a pretest to introductory biology students (n = 1396), as a posttest to other introductory biology students (n = 235), and as a posttest to intermediate-level students (n = 102). (A) Each whole question, scored as correct or incorrect (24 questions with three to five response options each). (B) Each response option, scored independently as correct or incorrect (92 T/F options). The dotted line indicates the hypothetical guess rate (50%).
Scores indicated that most questions were fairly difficult for the population tested. This is not surprising, given our design, which tests multiple levels of understanding within each question—most items require a fairly sophisticated conception of the central dogma for a subject to choose all the right responses and none of the distractors. Thus, we separated the data for introductory students tested preinstruction, whom we expect to have very weak conceptions, from the students tested postinstruction or in higher-level courses, whom we expect to have more expert-like thinking. The more advanced subset of students (N = 337) was used for the item-response analysis shown in Table 6.
| Question | P | D | rpb |
|---|---|---|---|
| Q1 | 0.31 | 0.56 | 0.57 |
| Q2 | 0.39 | 0.67 | 0.61 |
| Q3 | 0.39 | 0.21 | 0.30 |
| Q4 | 0.38 | −0.15 | −0.01 |
| Q5 | 0.09 | 0.05 | 0.19 |
| Q6 | 0.35 | 0.38 | 0.47 |
| Q7 | 0.46 | 0.43 | 0.49 |
| Q8 | 0.26 | 0.32 | 0.40 |
| Q9 | 0.12 | 0.33 | 0.52 |
| Q10 | 0.38 | 0.47 | 0.51 |
| Q11 | 0.51 | 0.53 | 0.52 |
| Q12 | 0.25 | 0.15 | 0.27 |
| Q13 | 0.04 | 0.06 | 0.20 |
| Q14 | 0.26 | 0.35 | 0.45 |
| Q15 | 0.36 | 0.62 | 0.58 |
| Q16 | 0.49 | 0.37 | 0.41 |
| Q17 | 0.22 | 0.25 | 0.36 |
| Q18 | 0.51 | 0.56 | 0.56 |
| Q19 | 0.47 | 0.60 | 0.59 |
| Q20 | 0.43 | 0.36 | 0.37 |
| Q21 | 0.64 | 0.41 | 0.43 |
| Q22 | 0.66 | 0.33 | 0.41 |
| Q23 | 0.18 | 0.23 | 0.39 |
| Q24 | 0.42 | 0.49 | 0.48 |
| Average | 0.36 | 0.36 | 0.42 |
The first analysis used all whole questions scored dichotomously, as correct or incorrect, and the difficulty index was calculated for each question (P = number of students with fully correct answer/all students with valid answers). Next, we considered each response option individually, wherein all 92 options were scored as correct or incorrect (Supplemental Table 1). Nearly all questions showed a positive correlation between item score and overall score (P), a positive discrimination value (D), and a positive point biserial correlation (rpb). One question, Q4, stood out as having negative D and rpb values. Even when looking at individual responses, it shows an unusual pattern: response A alone gives a low positive D and a low positive rpb, while choices B and C are negative in both respects. Questions 5, 13, and 23 each showed lower than ideal P, D, and rpb values, but all other questions appeared to fall within acceptable ranges. Cronbach’s alpha was calculated as 0.80 for version 4, and with the removal of Q4 it improved to 0.83, well above the threshold of 0.70 that is considered acceptable for a reliable instrument (Kline, 2000) and on a par with other assessments for college students (Liu et al., 2012).
Pre/Posttesting of a Molecular Biology Class
The above evaluation represents a snapshot of student performance at particular levels, in which students in different courses were evaluated at the same time. We also examined the change in performance of a single class over time. This course was particularly suited for the instrument, because it was an intermediate-level course that focused on concepts of cellular and molecular biology, with a particular emphasis on the central dogma. Student scores are shown in Figure 4; students in this class improved significantly from the pretest (n = 58) at week 1 to the posttest (n = 53) at week 14, t(109) = 5.47, p < 0.0001. Because the CDCI was given anonymously, it was not possible to correlate individual scores. Because five fewer students completed the posttest, we removed the lowest five scores from the pretest to give the most conservative estimate of the difference between pre/postscores. Removing the lowest five scores did not change our conclusion, t(104) = 4.8, p < 0.0001.
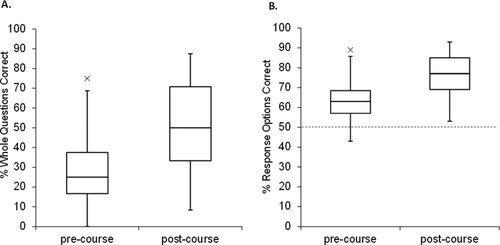
Figure 4. Results of pre/posttesting in a molecular biology class (CDCI v.4). The CDCI was given as a pre (n = 58) and post (n = 53) course assessment in a semester-long molecular biology course. The pretest was administered in week 1 and the posttest was administered in week 14. (A) Each whole question, scored as fully correct or incorrect (24 questions with three to five response options each). (B) Each response option, scored independently as correct or incorrect (92 T/F options). The dotted line indicates the hypothetical guess rate (50%).
DISCUSSION
We have developed a novel assessment tool designed to measure student thinking about topics related to the central dogma of molecular biology. Information flow, one of the five overarching core concepts articulated by Vision and Change (AAAS, 2011), is difficult for many biology students and is the subject of much biology education research (e.g., Pashley, 1985; Lewis and Wood-Robinson, 2000; Lewis et al., 2000; Marbach-Ad, 2001; Newman et al., 2012). The CDCI tool was developed by first examining student-generated ideas from research and classroom-based open-ended written assessments, formal research interviews, and informal interactions with students and biology instructors. Then items were designed, tested, and revised in an iterative process involving group discussions, student testers, and expert reviewers. We have provided evidence that supports the relationship between CDCI score and knowledge about topics related to the central dogma; the CDCI assessment can be used to generate valid inferences about a population of students. In this paper, we have described our instrument development and validation processes to the biology education research community in order to share our insights and experiences with instrument development to date.
Strategic Choices
While it is generally accepted that well-designed assessment tools rely on student-derived or novice ideas and language, it is nearly impossible to use student-derived language directly. As in other education research fields, our analyses of written artifacts from a number of biology students underscored the imprecise and vague nature of student language (Kaplan et al., 2009; Peterson and Leatham, 2009; Haudek et al., 2012; Rector et al., 2012; Wright et al., 2014). Many times it was difficult to figure out what exactly the student was trying to articulate when answering any number of question prompts. When designing CDCI items based on student reasoning and language, we recognized that we would need to transform vague novice language into specific item choices. For example, the term “convert” was used in many student-generated written responses attempting to describe the process of transcription, as in “The DNA is converted to RNA,” or “The T [thymine] is converted to U [uracil].” There are a number of possible interpretations of written statements that use the term “convert.” For example, did the student literally mean a chemical or physical conversion from one molecule to another? Perhaps the student thought part of a molecule was swapped out for something else? Or did the student actually understand the process of transcription but could not accurately describe what he or she meant? Because of this kind of ambiguity, raw student language could not be used verbatim in the assessment tool, but qualitative one-on-one interviews were helpful in uncovering underlying student ideas (Bowen, 1994; Otero and Harlow, 2009) and allowed us to better interpret and use student-generated language in the instrument. Thus, we attempted to decode student language from as many oral and written sources as possible in order to capture a wide range of student thinking. This process helped us create a number of possible item choices, both distractors and correct answers, for each question. The multiple-select format of the instrument provided us the freedom to use various combinations of correct and incorrect choices for each item. The extensive testing showed that every distractor is chosen as a “correct” option by at least a subpopulation of test takers.
In addition to allowing for more than one correct answer to be selected for each question, multiple-select format prevents users from utilizing “test-taking” strategies to correctly guess one correct answer found in a forced multiple-choice format (Haladyna and Downing, 1989; Towns, 2014). Well-designed items and responses, of course, greatly diminish the user’s ability to “guess” the correct answer, but they do not eliminate the problem of being able to narrow down the possibilities and then guess, which can inflate the chances of choosing the correct answer. Multiple-select items require that each response be evaluated independently. Thus, the multiple-select format is more likely to provide valid inferences without construct-irrelevant variance (Messick, 1995). Also, forced multiple choice does not help an instructor or researcher uncover alternative ideas the student may hold in regard to a particular question or topic (Couch et al., 2015a). For example, if a student is wavering between choices “A” and “C” on a forced-choice test, they will eventually have to choose one response, when in reality, the student might actually think that both “A” and “C” are correct. We feel a multiple-select format will help biology instructors and education researchers get a more accurate and complete picture of student thinking.
The statistical analyses of the beta test (version 4) helped us identify one question that needed to be discarded (Q4), and a few others that needed some rewording (Q5, Q13, and Q23). As shown in Table 6, questions were difficult in general (mean P = 0.36), as expected for a multiple-select instrument, with the difficulty index (P) spanning a range of moderately difficult (66% of students answering correctly) to very difficult (4% of students answering correctly). When considering all possible responses within each question, the ranges became broader (20–94%; Supplemental Table 1), as expected. The point biserial correlation (rpb) fell between 0.2 and 0.8 for 22 of 24 questions, with a mean of 0.42, which indicates that performance on individual questions is generally consistent with performance on the whole CDCI (Ding et al., 2006). The range of discrimination ability (D) for all questions ranged from −0.15 to 0.67, with only one being negative (Q4, which was removed for version 5) and 17 of 24 above 0.3. Thus, although it is not necessary to have all questions fall within the ideal range of 0.3–0.9 (Ding et al., 2006), most questions of the CDCI do fall within that range, and with a mean D of 0.36, the instrument overall discriminates reasonably well between top and bottom performers in our sample. A broad range of values is consistent with other recent non forced-choice biology assessment tools (Kalas et al., 2013; Couch et al., 2015a). Further information is gained by examining performance on individual options within items that have low D values, as many of the individual options do have the ability to discriminate more clearly between top and bottom performers (see Supplemental Table 1). This suggests that evaluating the specific responses that students chose via multiple-select format allows for a more accurate assessment of student understanding. For example, Q5 asks students to identify processes in which a nucleic acid exhibits catalytic properties. Looking at the expanded results (Supplemental Table 1), we can see that most students are aware that DNA synthesis and RNA synthesis are not correct answers, while protein synthesis is correct (the options are relatively easy to get right, and they do not correlate with overall score or discriminate between top and bottom). Far fewer students, though, correctly identify RNA splicing as a correct answer (P is much lower, while D and rpb are higher). This option addresses an advanced concept about molecular interactions that is not typically explained at an introductory level, so it is not surprising that very few students know it. Thus, specific responses to many of the questions, including Q5, expand the utility of the CDCI to be applicable to even advanced biology students.
Construction of an assessment tool involves a multipronged approach requiring the researcher to pay attention to many different issues. In addition to considering student conceptions of the content we wished to assess, we applied advice from the literature on differences in expert–novice reasoning strategies, inaccuracies of student-derived language, and assessment item–construction considerations. We made the choice, for example, to not include any graphical representations on the instrument, because that would have added an additional layer of cognitive load for students to decipher during the test. We present the development of the CDCI as a novel body of work, because we feel our strategies and methodologies may be helpful for others seeking to construct an assessment tool.
Scoring Considerations
While we feel a multiple-select format has many advantages, we must also highlight the challenges we faced with this format. First of all, not all automated scanning systems can grade multiple responses or parse out the partially correct answers from the completely wrong ones. Some software can grade only multiple-choice questions with one correct response per question; this type of software cannot identify patterns of responses or partially correct answers (e.g., “A” and “B” were chosen when “A,” “B,” and “E” were correct). An online format would make scoring multiple-select questions easier, but instructors would have to find ways of ensuring that students do not use outside sources to determine their answers, collaborate on answering test questions, or post the items on the Internet for others to consider. As with standard multiple-choice instruments, instructors and researchers are usually very interested in the patterns of incorrect answer choices made by their students or research subjects, not just the percentage who chose the right answer. Analysis of the data from a multiple-select instrument takes a little more time to gather and interpret, because there are many more patterns of student answers to analyze.
Scoring multiple-select items is also somewhat more complex than scoring a standard multiple-choice instrument. One way to score the CDCI is to consider each question as a whole and score it as completely correct (i.e., “A,” “B,” and “E”) or completely wrong (i.e., any pattern other than “A,” “B,” and “E”), which ignores partially correct answers and leads to lower scores (Albanese and Sabers, 1988; Tsai and Suen, 1993; Kalas et al., 2013). Another strategy involves grading each response individually, similar to a multiple true/false (T/F) format, which gives differential weight to different questions/concepts (since not all questions had the same number of options). The third way to score each item would be to assign a percentage of correct responses per whole question, wherein each option is considered against whether it should or should not have been marked to be correct. One consideration for our instrument is that not all questions in the CDCI have the same baseline guess rate; a question with one right answer out of three possible choices is different from a question with two right answers out of five possible choices. If answered randomly, the chances of marking anything other than the right answer is lower for the first case than the second (i.e., choosing the correct answer at random has a baseline guess rate of 1/7 when there are three options, 1/15 when there are four options, and 1/31 when there are five options). We chose not to force the same number of responses for all questions in order to preserve the authenticity of the items, because including artificial responses that are clearly wrong increases cognitive load for no worthwhile reason (Haladyna et al., 2002). Despite the challenges, every individual response option for every question is chosen by at least a small population of students. This suggests that the CDCI captures student thinking well and does not contain options that do not resonate with our test populations. The CDCI, at this point, is not a criterion-referenced assessment; we do not have enough data to support that a particular score is indicative of a particular skill level. The CDCI is a norm-referenced assessment tool; it can be used to identify performance in relation to the performance of other test takers (Popham and Husek, 1969).
Limitations
Many recent educational reforms (e.g., Cooper and Klymkowsky, 2013; Gottesman and Hoskins, 2013; Couch et al., 2015b; Dolan and Collins, 2015) have focused on strategies that encourage students to employ higher-level cognitive skills such as synthesizing and evaluating problems, rather than lower-level skills like remembering and applying (Anderson and Krathwohl, 2001; Crowe et al., 2008). While it is useful for instructors to use assessment questions at a variety of cognitive levels for formative and summative classroom assessments, the CDCI tool is designed to assess how well students grasp the fundamental concepts of central dogma topics. Higher Bloom’s level questions are valuable for assessing how well students can apply these foundational concepts to new scenarios. Using such questions on a concept inventory, however, does not allow an instructor to easily disentangle the reasons for incorrect response (i.e., did the student not understand the concept or did he or she not know how to apply the concept?). Examination of the difficulty index values reveals that this is a very challenging assessment, especially for first-year students. In fact, whole question scores are extremely low for new learners, because they have not learned enough to answer all parts of many questions correctly (e.g., in Q1, many lower-level students recognize that double-stranded DNA is needed for transcription but not the free ribonucleotides). Fortunately, the multiple-select design of the instrument allows instructors to identify which facets of the broad topics are most problematic for any population.
While our intent was to design multiple-select questions that did not contain any mutually exclusive choices, it was very difficult to adhere to this rule 100% of the time. Three questions (Q10, Q15, and Q17) contain pairs of item choices that are mutually exclusive. We acknowledge that the lack of independence could slightly impact inferences based on our statistical results, but we feel these questions are worth keeping because of the information provided by students’ answer choices.
As with most studies that utilize pre/posttesting, we cannot know the impact that repeated instrument exposure has on posttest performance. We strove to minimize this impact by spacing pre/posttesting as far apart as possible (14 wk) and by not discussing any CDCI question. Additionally, there was no course credit given for CDCI score, so students had little incentive to investigate the correct answers.
Finally, although we attempted to collect data from a wide range of demographics (institution size and type, class size and type), students differ widely in their prior experiences. We cannot say definitively that use of the CDCI would have similar results at all institutions.
Applications
The results presented here strongly support the value of the CDCI in its current format (version 5). We feel the CDCI will be a valuable tool to biology instructors and education researchers as a pretest to gauge student preparation, as pre/postassessment tools to measure pedagogical interventions, and possibly as a longitudinal instrument to test curricula. This type of instrument also may be useful for those in the research community who are interested in articulating learning progressions, descriptions of increasingly sophisticated ways of thinking about a topic (National Research Council, 2007). Learning progressions could potentially help coordinate instructor training, assessment strategies, and classroom activities centered around improving learning about information flow (Alonzo and Gotwals, 2012). Individuals who are interested in using the CDCI may contact the corresponding author for more information on how to obtain and implement the tool.
ACKNOWLEDGMENTS
This work was supported by a Genetics Education Research Program grant from the American Society of Human Genetics. We thank Dr. Thomas Kim for suggesting the multiple-select format and Dr. Brian Couch for help with statistical issues for this kind of instrument. We are grateful to the Science and Mathematics Education Research Collaborative at the Rochester Institute of Technology, the Biology Education Research group, Dr. Ross Nehm, and three anonymous reviewers for insightful suggestions. We also thank the faculty who helped evaluate the CDCI and/or administered it to their classes.



