Development of a Lac Operon Concept Inventory (LOCI)
Abstract
Concept inventories (CIs) are valuable tools for educators that assess student achievement and identify misconceptions held by students. Results of student responses can be used to adjust or develop new instructional methods for a given topic. The regulation of gene expression in both prokaryotes and eukaryotes is an important concept in genetics and one that is particularly challenging for undergraduate students. As part of a larger study examining instructional methods related to gene regulation, the authors developed a 12-item CI assessing student knowledge of the lac operon. Using an established protocol, the authors wrote open-ended questions and conducted in-class testing with undergraduate microbiology and genetics students to discover common errors made by students about the lac operon and to determine aspects of item validity. Using these results, we constructed a 12-item multiple-choice lac operon CI called the Lac Operon Concept Inventory (LOCI), The LOCI was reviewed by two experts in the field for content validity. The LOCI underwent item analysis and was assessed for reliability with a sample of undergraduate genetics students (n = 115). The data obtained were found to be valid and reliable (coefficient alpha = 0.994) with adequate discriminatory power and item difficulty.
INTRODUCTION
In 2009, the American Association for the Advancement of Science (AAAS) with support from the National Science Foundation published a report entitled Vision and Change in Undergraduate Biology Education: A Call to Action (AAAS, 2011). Vision and Change advocates for curricula that use facts to promote a deeper understanding of core biological concepts rather than curricula that promote cataloging of facts. (AAAS, 2011). Furthermore, it advocates for student-centered teaching methods in which instructors actively follow students’ progress in the practice of science rather than in the cataloging of facts (AAAS, 2011). Applying essential knowledge in a holistic manner to answer questions is more aligned with the modern practice of science.
Many core biological concepts are found in the domain of genetics, including evolution, inheritance, and the function of genes (Banet and Ayuso, 2000; Brownell et al., 2014). These core concepts have implications at the biological scales of the cell, organism, and population and are therefore applicable to all subdisciplines within biology. Despite their importance and prevalence in the field, genetics concepts are generally difficult for undergraduates to learn due to the highly technical language involved, the necessity of quantitative fluency, the requirement to think across several spatial scales, and the need to understand processes that are complex and often unobservable to the human eye (Knippels, 2002; Tibell and Rundgren, 2010; Karagoz and Cakir, 2011; McElhinny et al., 2014).
The abstract and sometimes foreign or counterintuitive nature of genetics subject matter lends itself to the production of conceptual errors (Browning and Lehman, 1998; Venville and Treagust, 1998; Banet and Ayuso, 2000) also known as misconceptions. Considerable work has been done on misconceptions in some areas of biology, particularly at the secondary school level, in the areas of genes and inheritance (e.g., Venville and Treagust, 1998; Wood-Robinson et al., 2000; Lewis, 2004; Gericke and Hagberg, 2007) and evolution (e.g., Bishop and Anderson, 1990; Greene, 1990; Anderson et al., 2002; Nehm and Reilly, 2007; Andrews et al., 2012), both of which are central to genetic and biological understanding. Relatively fewer research studies, again primarily at the secondary school level, have been done on concepts regarding more molecular topics, such as, the relationship between genotype and phenotype, the chemical origins of mutations, and the effects of mutations on organisms (Smith et al., 2008; DeHoff, 2010; Todd and Kenyon, 2015).
Identifying and acknowledging student misconceptions is important, because an understanding of biological sciences is increasingly important for society, as it informs views on healthcare, educational curricula, climate change, and food sources. Together with nationwide calls for more graduates with degrees in science, technology, engineering, and math, there is also an imperative need for all citizens to be at least biologically literate (President’s Council of Advisors on Science and Technology, 2012). Students, who are both citizens and possible future scientists, benefit from our understanding of conceptual errors, because such errors inform teaching practices. Existing instructional methods could be amended or new methods could be created, used, and assessed to aid in learning (Engelman and Huntoon, 2011).
In the rapidly expanding field of genetics, modern genetics increasingly builds on the foundational understanding of core concepts. Gene regulation is one such concept because it is applicable to many higher-level genetics and biology concepts and hands-on laboratory techniques. Additionally, it is a core principle for the complex, interacting systems used in biomedical, environmental, and pharmaceutical research and development endeavors. For example, human insulin, which is used by diabetics, is currently produced in bacteria using bacterial gene regulatory regions in control of the human insulin gene (Walsh, 2005). Bioremediation of nuclear waste sites has been enhanced by genetic engineering of a uranium-degrading bacterium called Geobacter sulfurreducens (Cologgi et al., 2014). For these reasons, gene regulation has been identified as one topic crucial to meeting the goals established by the nationwide calls for reform in undergraduate biology education suggested by Vision and Change (AAAS, 2011; Brownell et al., 2014). However, many students are known to hold common misconceptions regarding gene regulation. The naïve and inaccurate model is that any gene that is present is expressed and that if a gene is not expressed, then it is not present (Bowling et al., 2008; Smith et al., 2008).
Gene regulation is actually achieved by a wide range of mechanisms that cells use to control whether or not genes are transcribed and when genes are transcribed and to increase or decrease the quantity of certain proteins based on the cellular and/or environmental feedback. One of the best-understood examples of gene regulation is the negative inducible lac operon of Escherichia coli (Figure 1). Regulation of the lac operon was first described by François Jacob and Jacques Monod. Their research demonstrated how enzyme quantities can be controlled directly at the level of transcription (Jacob and Monod, 1961). Their discoveries gave rise to a large subdiscipline within molecular biology devoted to the understanding of genetic regulation and have become well established in the curricula of undergraduate genetics and microbiology courses.
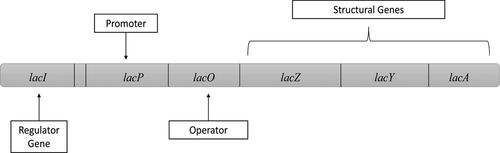
Figure 1. The lac operon. A basic schematic of the operon and its regulatory elements. A version of this was included on the LOCI for students.
Two well-understood model transcriptional regulatory systems for learning the basic principles of gene regulation are the trp and lac operons, one of which is inducible and the other repressible. Having a firm understanding of one of these operons makes understanding the other simpler, as their mechanisms of action only vary in how binding at the allosteric site affects the repressor. Once students have a grasp of these examples of prokaryotic gene regulation, that knowledge serves as a base for the scaffolding of other bacterial operons and also more complicated systems of gene regulation, such as complex eukaryotic transcriptional regulation, epigenetics, negative-feedback loops, molecular cloning, and systems biology (e.g., Cronan et al., 1998; Olaharski et al., 2005; Zoller et al., 2015). While the focus of this particular CI is a single operon, instruction within the context of the entire course would provide opportunities for students to observe and evaluate the similarities and differences in other gene regulation systems.
Given the importance of this topic and evidence showing that students struggle with genetics concepts that go beyond rote learning and require critical thinking, our larger study was driven by an investigation of better ways to teach and assess students on the lac operon (Cavallo, 1996; Lewis and Wood-Robinson, 2000; Lewis, 2004). It is critical that learning methods be empirically evaluated to determine their efficacy. Assessment tools are needed for these evaluations and to aid in identifying student misconceptions to inform teaching practices (Knight, 2010). These tools, also outlined in Vision and Change, can come in many forms, ranging in their ease of implementation, their efficacy in accurately capturing student understanding, and their reliability. They include true/false questions, one-minute papers, concept maps, concept inventories (CIs), and research papers synthesized by students (AAAS, 2011).
Owing to their utility, a growing body of CIs, ranging from broad general introductory biology inventories to more narrow inventories dealing with specific concepts such as meiosis, has been published (D’Avanzo, 2008; Kalas et al., 2013). A CI is relatively simple to administer to large numbers of students, and the data provided by a CI are easily quantified and analyzed (Knight, 2010). These factors make CIs useful and reliable tools for assessing what students know and common misconceptions they may retain. In fact, in her 2013 follow-up to Vision and Change, D’Avanzo (2013) suggests that a body of CIs be used as a means of providing data as evidence to biology department faculty members to confirm the value and support the use of active learning and student-centered course design. An emphasis was placed on the importance that active-learning exercises be carefully designed for specific concepts within a course and not just for the course overall (D’Avanzo, 2013).
As part of our larger research project of developing undergraduate learning tools for specific concepts in prokaryotic gene regulation, we searched for a valid and reliable tool to measure student learning regarding gene regulation with little success. For example, the Genetics Concept Assessment developed by Smith et al. (2008) does not include items covering gene regulation. The Molecular Biology Capstone Assessment has three of 18 questions on gene regulation, only one of which is relevant in prokaryotes (Couch et al., 2015). More recently, highly specific CIs have been developed and validated for other specific genetics concepts, such as the Meiosis Concept Inventory (Kalas et al., 2013), the Transcription/Translation Concept Inventory (Questions for Biology, 2015), the Dominance Concept Inventory (Abraham et al., 2014), and the Genetic Drift Concept Inventory (Price et al., 2014). Through our search, we located no existing CI that assesses basic knowledge of gene regulation by testing understanding of the lac operon. As such, our pragmatic need to assess student learning became an opportunity for instrument design. Using a rigorous protocol for CI design (D’Avanzo, 2008; Knight, 2010; Adams and Wieman, 2011; Kalas et al., 2013), we have constructed and validated a CI and have identified some common misconceptions related to aspects of the lac operon regulatory system.
While valuable information can be gained by looking at individual scores on the Lac Operon Concept Inventory (LOCI), group performance would be important for assessing effectiveness of particular teaching strategies, documenting the persistence of misconceptions, and assessing shifts in expert-like thinking. In addition, while the LOCI was designed to examine four conceptual areas of prokaryotic gene regulation, the first three concepts are more commonly taught in microbiology, so it may be of use to instructors of microbiology or biotechnology courses as well as in genetics courses. It is hoped that the availability of this CI will remove hurdles for faculty members who wish to assess and perhaps publish their own teaching methods and activities regarding gene regulation but do not have the time to develop such assessments of their own. Well-designed CIs allow for valid and reliable means to assess student knowledge of particular concepts that may not be captured in other more traditional assessments. As such, the fine-grained analysis of particular items allows educators to adjust instruction tied to particular learning objectives and enhance the overall learning experience for students. The authors are currently using this CI as a tool to assess an active-learning module on prokaryotic gene regulation for undergraduate genetics students.
METHODS
Study Participants
This study was conducted between May 2014 and December 2014 at a large public university in the southeast United States with more than 22,000 students. The student population is 54% female and has an average ACT score of 22.3. Participants were registered in undergraduate microbiology or genetics courses. Demographic data were not collected from individual participants. Combined, these courses enroll more than 1000 students per year. Microbiology students at this institution are typically freshmen or sophomores, while genetics students are typically sophomores or above. This study was conducted using ethical protocols for using human subjects and was approved by the Internal Review Board at MTSU (IRB 15-025). Only data from those students who gave their informed consent were included in this study. As different numbers of students participated in different aspects of the inventory-design process, the sample sizes for each stage are indicatedin in Table 1.
| Step | Purpose | Sample size (n) | Feedback type |
|---|---|---|---|
| 1. Learning objectives established and initial questions developed (Spring 2014) | Expert review of questions | ||
| 2. Piloting first iteration (May semester and Summer session 1, 2014) | Identify misconceptions (distractors) and clarity of questions | 22, 18 | Short written responses, class discussion |
| 3. Piloting second, multiple-choice iteration (Summer session 2, 2014) | Wording/phrasing of questions and answer choices | 23 | One-on-one interviews |
| 4. In-class testing (Fall 2014) | Item analysis, validity, and reliability | 100, 15 | In-class administration of 12 multiple-choice questions |
Inventory Design
Our larger study involved testing the utility of a hands-on model for learning regulation of the lac operon. The intended group for this activity is college students enrolled in genetics, microbiology, and biotechnology courses or any other course in which they learn the lac operon regulatory system. The goal was to design an instrument that would be used to compare conceptual understanding between groups and that would reliably provide accurate data on those conceptions. Our LOCI was created following established, peer-reviewed procedures for CI design (D’Avanzo, 2008; Knight, 2010; Adams and Wieman, 2011; Kalas et al., 2013; see Table 1). The research team consisted of a faculty member with expertise in genetics and 15 yr teaching experience in undergraduate genetics (R.L.S.-T.), a graduate student with expertise in both genetics and education (K.M.S.), and a faculty member with expertise in biology education research (G.E.G.). First, using a backward instructional design (Wiggins and McTighe, 2005), the following student learning objectives were agreed upon by the research team and contributed to the initial content validity of the LOCI. Following instruction, students were expected to be able to
identify and understand the role of the structure and components of the lac operon;
when given particular cellular conditions, accurately predict whether or not gene expression will occur; and
when given particular mutations to the lac operon, predict affected outcomes of gene expression.
It was agreed among the authors that these learning objectives were appropriate for the target learners. With the learning objectives in mind, and using items from previous educational resources as a guide, we carefully constructed 12 open-ended short-answer pilot inventory questions aligned with our learning objectives. The authors wrote items that they felt adequately covered the nuance of each conceptual learning objective. The learning objectives vary in complexity, so the authors did not expect each objective to have the same number of items. The number of items was a balance between 1) constructing an overly long assessment for pragmatic and validity reasons and 2) the minimum the authors felt necessary to observe conceptual understanding based on teaching experience in the area. Four items were written for the first learning objective and two for the second. Six questions were written for the third learning objective, which requires a complete understanding of the first two objectives. These initial open-ended items were then reviewed by three biology department faculty members (who each hold a doctoral degree in either microbiology or molecular biology and who have taught the lac operon) to ensure readability, representativeness, appropriate difficulty level, and content aligned with the learning objectives. Students in both microbiology and genetics courses were involved in the CI design and validation to allow a more timely design process. Both the microbiology and genetics courses are commonly taken in the sophomore year at this institution, and both courses include instruction on the lac operon, so these students would be expected to represent similar current and prior knowledge levels. The open-ended item set was initially given to a section of microbiology students (n = 22) as a means to catalogue common student misconceptions related to the learning objectives. Students were instructed to answer the items in writing with as much detail as possible. In addition, they were asked to provide feedback on the items themselves to help clarify the wording. This was done to give students the opportunity to provide open, honest responses and feedback on both the concepts and the items themselves. Students answered the pilot inventory immediately following their in-class lecture on the topic. Following administration of the pilot inventory, the items were discussed with the class as a group to receive additional verbal feedback regarding clarity of the item wording. Written feedback at this point was inadequate, as only a small number of students completed the inventory fully and as directed. While the feedback was not sufficient at this point to gain a full picture of students’ misconceptions, two issues with the wording of the short-answer items were identified in the verbal feedback session. Students were confused by the term “polycistronic” and indicated that more detailed instructions would be helpful when answering the questions about mutations.
Following reflection, the researchers elected to obtain more student feedback, because the feedback obtained from this first pilot was sparse and incomplete. Gathering additional data would necessarily improve the content validity and readability of the items and would identify misconceptions. As such, the pilot inventory was then given to a second group of students. This pilot iteration was almost identical to the first iteration, with the exceptions of removal of the term “polycistronic” and more detailed instructions for the items pertaining to mutations. These items were administered, with the same instructions with respect to providing written responses and feedback, to a second microbiology class of 18 students with the added incentive of extra credit for thoroughly answering each item. These students had attended a lecture on the lac operon the previous day. The added incentive was enough to ensure sufficient feedback was provided on the items, as they were completed much more comprehensively.
Students’ responses were carefully read through to identify any misconceptions held by students on each open-ended item. These short-answer responses were analyzed to identify areas in which students had difficulty and made mistakes. Some responses indicated a complete lack of understanding of the concept and had to be disregarded. However, many responses showed that students had some level of understanding of the lac operon but demonstrated errors in their understanding of specific details or relationships within the topic. Through content analysis, it was from these responses that we began to identify common misconceptions and students’ preferences for word choice.
Using these misconceptions from the first two groups of students on the first two pilot iterations of the CI, we drafted 12 multiple-choice items that aligned with the established learning objectives. Word choices used by the students were identified and used when constructing the items and answer choices. For example, we use the term “promoter” as an answer choice for item 1 instead of lacP. Common errors identified in the pilot iterations, student discussions, and student interviews were used as distractors. Please see Results for details.
To improve the validity of the data obtained from the assessment, the draft multiple-choice LOCI was reviewed by one faculty expert and administered to students from a genetics course (n = 23). We followed an administration procedure similar to that used by Kalas et al. (2013) in their design of a meiosis CI. The first author met with individual students outside class and had them answer the LOCI verbally while talking out their reasoning as they answered each item. Generally, students indicated that the items were clear and representative of what they had learned. Using student feedback the wording of item 5 was adjusted from “Is mRNA transcribed from the lac operon when lactose is present in the cell? Why or why not?” to “Is mRNA transcribed from the lac operon when lactose is present AND glucose is not present in the cell? Why or why not?” At this stage in the process, many of the same common misconceptions were once again evident, providing convergent validity to the misconceptions identified and indicating that saturation was likely.
Statistical Analysis
Finally, the LOCI was reviewed by two expert faculty members, both of whom hold doctoral degrees and have expertise in genetics and experience teaching the lac operon. They agreed with the authors on the correct answer for each LOCI item and agreed the content was appropriate for the target student group. The LOCI was then administered to a larger sample of genetics students (n = 115) from two different genetics courses taught by two different instructors. Students’ responses were used to create a frequency distribution for each item showing how often each answer choice was selected. This allowed us to identify the most commonly selected answer and commonly selected distractors.
Item analysis, including index of difficulty, item discrimination index, and point-biserial correlation (Table 2), was conducted as previously described (Findley, 1956; Doran 1980). The index of difficulty (P) was calculated by finding the proportion of students who gave the correct answer for each item. The index of difficulty indicates how challenging a particular item is in this context and ranges from 0.00 (very easy item) to 1.00 (very difficult item). Experts suggest a range of 0.60–0.80 as optimal when constructing multiple-choice items (Kubiszyn and Borich, 2003). The discrimination index and the point-biserial correlation both compare a student’s score for an individual item with how well he or she performed on the overall assessment. Item discrimination index values range from −1.0 to +1.0. A value of +1.0 would indicate that every student in the top-performing group answered the item correctly and every student in the bottom group answered incorrectly, whereas a value of −1.0 would indicate that every student in the bottom-performing group answered the item correctly, while every student in the top group answered incorrectly. A point-biserial correlation is a measure of reliability for each item on the inventory. The values for correlation range between −1.0 and +1.0. Point-biserial correlations will be positive if students with higher total scores are more likely to answer the question correctly than students with lower total scores. Both of these indices indicate whether an item (or assessment) is appropriately differentiating between low- and high-knowledge students. This should be the goal of a well-constructed assessment item. Additionally, a coefficient alpha was calculated using IBM SPSS Statistics 21 software.
| Learning objective | Item | Sample size (n) | Index of difficultya | Discrimination indexb | Point-biserial correlationc |
|---|---|---|---|---|---|
| Knowledge of operon structure and its components | 1 | 115 | 0.42 | 0.19 | 0.36 |
| 2 | 115 | 0.77 | 0.42 | 0.44 | |
| 3 | 115 | 0.55 | 0.12 | 0.32 | |
| 4 | 115 | 0.40 | 0.48 | 0.43 | |
| Predicting outcomes of various cellular conditions | 5 | 115 | 0.46 | 0.45 | 0.34 |
| 6 | 115 | 0.34 | 0.48 | 0.40 | |
| Understanding the effects of known mutations | 7 | 109 | 0.54 | 0.65 | 0.54 |
| 8 | 113 | 0.42 | 0.56 | 0.53 | |
| 9 | 115 | 0.49 | 0.52 | 0.44 | |
| 10 | 114 | 0.30 | 0.23 | 0.32 | |
| 11 | 109 | 0.46 | 0.42 | 0.42 | |
| 12 | 109 | 0.50 | 0.55 | 0.47 | |
| Desired values | 0.3–0.9 | ≥0.3 | ≥0.2 |
Limitations
The authors acknowledge the limitations of this study. Data from this study were limited to a single university context, but the setting is large and is a reasonable representation of this population of students. Future studies using the LOCI might want to provide detailed demographic data allowing for cross-institutional comparison. Ideally, larger samples might have been used during the early iterations; however, both pilot iterations and the open-ended interviews included discussions with students that should have brought out common errors and misconceptions in a total of 63 students before the draft multiple-choice LOCI was developed. With the qualitative data and cross-validation with multiple experts, we are confident in our target objectives.
RESULTS
The LOCI was developed using rigorous methods established previously (D’Avanzo, 2008; Adams and Wieman, 2011; Knight, 2010; Kalas et al., 2013). To facilitate the use of the LOCI without allowing inappropriate student access, the final LOCI questions are available to instructors from the corresponding author.
Analysis of Student Thinking from Pilot Iterations
Two short-answer/group discussion pilot iterations of the inventory were conducted with a total of 40 students to investigate student thinking in response to short questions on the lac operon. As noted in Keeley (2012), misconceptions can vary by degree from factual to conceptual. More deeply entrenched conceptual misconceptions would be more difficult to correct and might require extensive discussion or activity, while mild, factual misconceptions might be corrected by pointing them out in a sentence or two. For this analysis, misconceptions were considered reasonable errors that were observed in student responses regardless of the degree.
The first four questions of the pilot assessment aligned with learning objective 1. In response to the item 1, “The lac repressor binds to what site within the lac operon?,” a small number of students answered “promoter,” “the lacP site” and “the regulator,” while a majority answered correctly. Both the promoter and lacP site answers are reasonable, because they are regions of DNA associated with the lac operon, but they are scientifically incorrect and could therefore be considered misconceptions. Students performed overwhelmingly well on item 2: “Is the gene for the lac repressor a structural gene in the operon? Does it encode for a protein that is directly involved in metabolizing lactose?” Students were able to identify that the lacI gene is not a structural gene contained within the lac operon. This question is relatively simple and was retained in the following multiple-choice iteration of the LOCI. In response to item 3, “The lac repressor is inactivated by binding to what molecule? Where does this molecule come from/what is it derived from?,” most students knew that the answers were “allolactose” and “lactose,” respectively. However, several students incorrectly answered, “The lac repressor is inactivated by binding to the operator.” This answer is also reasonable, in that the repressor does bind to the operator in other circumstances, but it is scientifically inaccurate in this case, and thus may also be considered a misconception. To item 4, “Which portions of the lac operon encode proteins that play a role in breaking down lactose and which play a role in controlling transcription?,” most students included lacP (promoter) and lacO (operator) as playing a role in regulation; however, several students also included lacI (regulator gene) in their answers. These answers fit into a broad category of misinterpreting the repressor gene as part of the operon (lacI), making their answers partially correct. The answers represent a misconception: while the function of lacI as a repressor (not a catalytic enzyme) is a part of how transcription is controlled, it is located in its own operon elsewhere in the genome and is therefore under the control of a distinct regulatory system with its own promoter separate from the other lac genes.
Items 5 and 6 addressed learning objective 2. Item 5 asked, “Is mRNA transcribed from the lac operon when lactose is present in the cell? Why or why not?” This item was frequently missed, and the following are examples of incorrect answers given by students that demonstrate a breakdown in their understanding of this process: “No, because the repressor will be inactivated by the inducer. The cell will be busy breaking down lactose instead of transcribing mRNA,” and “No, mRNA is not transcribed from the lac operon when lactose is present. It is not transcribed because when lactose is present there’s no allolactose to bind to the lac repressor.” The first student response, an incorrect answer of “no” followed by a scientifically correct answer of what binding is occurring, indicates a disconnect between the function the repressor and consequences of its binding allolactose, clearly a misconception. Interestingly, this response ends with a somewhat teleological justification statement, as in Coley and Tanner (2012, 2015), that implies the cell makes a choice to break down lactose rather than produce RNA. The second response shows the student has a much more easily corrected misconception—that is, allolactose is simply a metabolite of lactose. In response to question item 6, “Is the lac repressor active (bound to DNA) in the absence of lactose within the cell? Why or why not?,” several students said “no.” The following are examples of their reasoning: “No, the repressor is acting to keep metabolism from occurring. If there is no lactose present then there is nothing to repress,” and “The lac repressor is inactive i[n] the absence of lactose within the cell because lactose must be present to activate the repressor.” This indicated that, while students might have been able memorize the role allolactose and lactose play in relation to the repressor (item 3), they did not connect that the role of the active repressor is to prevent transcription, which in turn saves energy, even when the short-answer question clarified that the active repressor is bound to DNA.
Items 7 through 12 were aligned with the third learning objective. Many students struggled with these questions regarding mutations, because they were not specifically taught about the mutations in class. However, several students were able to use their understanding of the operon to work out the correct answers to these questions. Because the microbiology students were not lectured on mutations, these questions (7, 9, 11, and 12) contained concise descriptions of each mutation. Instructions for this section specifically asked students to answer in terms of whether or not transcription of the operon and protein synthesis would occur. Items 7 and 9 were followed up with a question (8 and 10) asking what would happen in each mutant if a wild-type copy of the gene were inserted into the cell (complementation). Item 7 stated, “A mutation known as lacI− in the lac repressor gene causes the repressor protein to be absent/nonfunctional. What effects would you expect to see in an E. coli cell that has this mutation under the following conditions: a. the presence of lactose? b. the absence of lactose?” The following is an example answer that demonstrated a conceptual error regarding the regulatory mechanism (available promoter = transcription occurs): “In the presence of lactose both transcription and protein synthesis could not take place. This is because the repressor is absent to begin with so the inducer could not bind. [In the absence of lactose] I don’t believe any transcription or protein synthesis would take place because again the repressor is absent.” The following is one response that demonstrated a misconception that was the reverse of the correct answer to item 8: “The repressor would be able to bind to the operator [in the presence of lactose]” and in the absence of lactose, “both the normal and gene and the mutated gene would not be regulatory at this time.” Some students were able to accurately determine that a wild-type copy of the gene would restore appropriate expression of the genes of the operon; however, multiple students indicated that the wild-type gene would “replace” the mutated gene, which appears to be a common naïve misconception. Item 9 described a lacIS, “superrepressor” mutation. Again, some students were able to correctly reason that this mutation causes the lac genes to never be transcribed regardless of the presence or absence of lactose. The following are examples of other students’ confusion: “It would continue to transcribe RNA because allolactose cannot bind to the repressor which causes transcription to continue in the presence of lactose,” and “In the presence of lactose I think that the lacIS would maybe be overlooked and the inducer would still bind to the repressor.” The first response may represent a misconception that occurred due to prior knowledge of a nonfunctional repressor, because the reasoning is consistent with the nonfunctional repressor mutation. The second response invokes special teleological circumstances of the cell “overlooking” its cellular environment and is thus a broader misconception that may not be related to gene regulation. Responses to the follow-up question about this mutation showed many misconceptions. The following are examples of answers given by three different students: “The insertion of a normal copy of lacI would lead to the normal production of a repressor. A normal repressor, in the presence of lactose binds lactose and thus unbinds from DNA and transcription occurs,” “If lactose is present then we would hope to see the mutated gene disposed of as junk then see the correct gene [transcribed],” and “Inserting a normal cop[y] of the lacI gene would allow for normal transcription leading to translation in both the presence and absence of lactose.” The reasoning in these answers is correct if the situation named has occurred, that is, if the mutant gene was replaced by the wild-type gene, but the replacement was an incorrect assumption students made based on some prior knowledge. Item 11 described the lacOC mutation. Interesting answers that demonstrated misconceptions include “Without lactose, proteins cannot be synthesized whether or not there is a mutation present,” and “In the presence of lactose the repressor would bind the inducer and start the process of transcription with this mutation.” Both responses indicate a misconception in the regulatory mechanism regarding the function of the operator in the system. Finally, item 12 described a loss-of-function mutation in the lac promoter. The following demonstrate students’ misunderstanding of the role of the promoter: “If lactose is present, I think transcription and protein synthesis will still occur,” and “I think the cell would function normally because the operator and repressor are unaffected by this mutation.” Both of these responses indicate a misconception dealing with the larger topic of transcription. Using these misunderstandings and misconceptions, we were able to construct the pilot multiple-choice version of the CI and determine the extent to which these misconceptions are common in this population of students.
Following the interviews with genetics students using the pilot multiple-choice version of the CI, some commonly held misconceptions were once again evident. Ten out of 23 students incorrectly answered that the repressor binds to the promoter. These students also struggled with understanding when the repressor is active. Eight students thought that the inducer activates the repressor, while seven accurately understood that repressor is active in the absence of the inducer. This provided convergent validity that common misconceptions identified in the open-ended items were also common in the LOCI items.
Item Analysis
A number of statistics were used to estimate the validity and reliability of the data that can be obtained using the LOCI. Index of difficulty, item discrimination index, point-biserial correlation, and the coefficient alpha were determined as described previously (Kalas et al., 2013). The index of difficulty is the proportion of students who select a correct answer and is used to determine whether particular items are appropriate for the group based on their current level of understanding of the topic. A widely accepted ideal range for difficulty index is 0.3–0.9. The rationale for this is that items within this range contribute to a test’s discriminability (Doran, 1980). For the LOCI, the difficulty index of one item was above 0.7, three fell between 0.5 and 0.7, and seven items fell between 0.3 and 0.5. This suggests that the items cover a range of difficulty with the majority falling into the moderately difficult category (Adams and Wieman, 2011; Kalas et al., 2013; Table 2).
The discrimination index and the point-biserial correlation measure the ability of an item to distinguish between high- and low-performing students. The discrimination index for the majority of the items on the LOCI was ≥ 0.3 (except items 1, 3, and 10), with a mean discrimination index of 0.42. The point-biserial coefficient of all items fell above the recommended value of ≥0.2 (Ding et al., 2006; Table 2). This indicates that most items have the ability to distinguish between high-performing students and low-performing students (Doran, 1980; Ding et al., 2006).
Finally, the coefficient alpha, which is mathematically the same as the Kuder-Richardson reliability index for this type of binomial (i.e., 0/1) data, is a measure of internal reliability or whole-test consistency. The coefficient alpha for the LOCI was found to be 0.994, suggesting a very high internal reliability for data gained. This value is extremely high, but given that the scope of the LOCI is quite narrow, it does not seem unreasonable. In addition, while values greater than 0.7 indicate reliability for group measurements, values greater than 0.8 are considered reliable for individual measurement (Doran, 1980; Ding et al., 2006). This value, therefore, suggests even greater utility for the LOCI in assessing individuals as well as groups.
Misconceptions Identified
One value of conducting rigorous item analysis and reliability testing on a CI is to ensure validity and reliability in identifying common misconceptions through student responses. Student misconceptions were first identified in the development of the LOCI through open-ended pilot items and interviews in which students talked through their answers, as discussed in the Methods. Next, frequent incorrect answers were then worded and used as distractors in the multiple-choice CI questions. The frequency with which these distractors are chosen is a good indicator of how many students hold a misconception for that topic, because the initial misconceptions were observed originating independently from several students through the course of student discussions and interviews.
Frequency distributions for each LOCI item, including the correct alternative and distractors, indicated several misconceptions in lac operon concepts that were indeed consistent from the pilot iteration forward (Figure 2). The most prominent of these was the mistaken belief that the lac repressor binds to the promoter, not the operator, as evidenced by students choosing this answer 37.4% of the time from item 1. The binding of the inhibitor to the operator, which is not the RNA polymerase binding site (promoter) itself, is an important issue, as it reveals information about the mechanism of gene regulation. Gene regulation in this instance is due to steric hindrance, a common but not exclusive mechanism of gene regulation. This mechanism is seen frequently in both prokaryotic and eukaryotic transcriptional regulation (McMurry and Levy, 2010; Holloway and Spirov, 2015) and is therefore an important introduction to the idea. Gene regulation that occurs by competition for the same binding site is a different but related mechanism commonly seen for transcription factors in eukaryotes (Hermsen et al., 2006). In addition, regulatory control that occurs due to sequences and circumstances unrelated to the RNA polymerase binding site (promoter) is also an important concept that is particularly true in eukaryotes, in which regulatory elements can extend several kilobases upstream, involve very complex differential regulation (Liu et al., 2011), and involve repression due to localized chromatin structure (Falvo et al., 2013). Further, the fact that the RNA polymerase binding site and operator are distinct and functionally different is important to understanding the effects of mutations, that is, why the operator mutation allows transcription to occur constitutively instead of stopping transcription altogether, as would be expected for a promoter mutation. One of the authors (R.L.S.-T.) has observed this misconception many times in her 15 yr experience of teaching prokaryotic gene regulation.
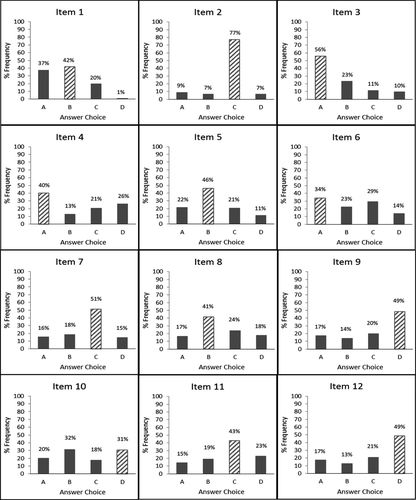
Figure 2. Frequency distributions for each item of the LOCI. The frequency of the correct answer choice is shown in hatched bars and the distractors in solid black bars from in-class testing (n = 115) of the LOCI. Data from items 1 and 10 both show commonly held misconceptions in this group of students, while item 6 shows a poorly understood concept. Data from item 2 indicate an idea that was well understood by this group of students.
The frequency distribution for item 6 showed that many students (29.6%) had a mistaken belief that the repressor is activated by the inducer. Another 22.6% of students incorrectly believed that lactose activates the repressor. This also is an important and far-reaching concept, as it relates functional consequences to protein–ligand interactions. This is seen in other prokaryotic gene regulation systems, such as in the repressible trp operon and also in eukaryotes, particularly in transcription factors. For example, the hormone estrogen binds its receptor, which is then translocated to the nucleus, where the protein–ligand complex binds estrogen response elements and activates transcription of those associated genes (reviewed in Klinge, 2001). This misconception has also been observed frequently in the same author’s experience, as it appears that students become easily confused by negative regulatory situations in general, but particularly when they are represented as “normal” occurrences and not as a result of some kind of mutation.
Item 10 was another poorly understood item, with 31.3% of students believing that introducing a wild-type copy of lacI into a cell with a superrepressor (lacIS) would result in a wild-type phenotype, while 30.4% correctly answered that it would not. One of the authors (R.L.S.-T.) suggests, through personal interactions with students, that this may indicate that students hold a mistaken belief that, when another gene is introduced into a cell on a plasmid, it replaces the existing gene. This concept is important from many different perspectives. Contained within the concept are several broader concepts: 1) the idea of multiple alleles (along with wild-type lacI and lacI−), 2) the biochemistry of dominance for a mutant allele (lacIs is dominant over both lacI and lacI−), and 3) the behavior of introduced DNA in a cell. The growth of genome sequencing has revealed that the two-allele system is a gross oversimplification for all genes, so this is a small introductory example of the idea. For example, the CFTR gene, which is the causative gene for cystic fibrosis, has more than 1200 nucleotide-level genetic variants and more than 200 structural variants (see ClinVar and dbVar at the National Center for Biotechnology Information website; www.ncbi.nlm.nih.gov). Allele dominance, an inherently important concept for understanding genotype–phenotype correlations in diploid cells, can be introduced with this system. For example, GLI3 mutations can cause both Greig cephalopolysyndactyly syndrome (GCPS; available at www.omim.org, unique identifier MIM#175700) and Pallister–Hall syndrome (PHS; available at www.omim.org, unique identifier MIM# 146510) in an autosomal dominant manner by a dominant negative mechanism for GCPS and a haploinsufficiency mechanism for PHS (Demurger et al., 2015).
In all, identification and knowledge of these misconceptions provide opportunities for educators to reconsider how they teach prokaryotic gene regulation and to develop better learning tools and activities that will allow students to confront and address their own misconceptions regarding prokaryotic gene regulation. The development and validation of the CI will allow assessment of these new strategies.
DISCUSSION
In recent years, CIs have been recognized for their value to educators as tools for assessing student learning and for informing instructional methods (Knight, 2010). Education research benefits from the increasing use of CIs, which provide a source of quantifiable data that clearly indicate student understanding and conceptual change. By creating an efficient means of categorizing student conceptions and misconceptions, CIs are a crucial part of the future of biology education research. With a repository of CIs available, researchers could adapt and apply them to studies that would result in powerful and valid inferences being drawn based on evidence.
Adding to an existing body of CIs, the LOCI provides an inventory that covers negative inducible gene expression. Because creating a CI is time and labor intensive, having the LOCI accessible provides a method for researchers and instructors to collect data that can be incredibly informative without necessitating that a CI be written and validated for each study. It can be used to examine students’ understanding of the lac operon and/or in the continued exploration of active-learning techniques applied to learning the lac operon. For example, in his 2015 paper, Robert Cooper outlined a comprehensive approach to teaching five “big idea” biology concepts with operon models (Cooper, 2015). His paper provides the ideas and activities but does not supply assessments to support the efficacy of the instructional approaches. The LOCI could be implemented in classes being taught as described to examine how well students understand the big ideas using operons, whether or not they have any widely held misconceptions, and what those misconceptions are. The results from the LOCI could then provide instructors with data to support or refute the use of Cooper’s approach, therefore potentially broadening the impact of his work. These types of data, from either CIs or other forms of assessments, are crucial to advancing the use of evidence-based, active-learning techniques and supporting education research and the broader discourse in the literature.
For the purposes of our larger study, we used the LOCI to compare treatment groups that had used a novel hands-on model to learn the lac operon with comparison groups who had learned the lac operon only through traditional lecture. While we did use the LOCI to collect prescores for these groups, we found that, because most students had no previous knowledge of the lac operon, any pre–post comparison would be inappropriate. As a result, we used the postintervention scores to compare conceptual understanding between groups. However, a pre–post comparison could be made in upper-division courses in which students are expected to build on previous knowledge of the lac operon when learning the regulatory mechanisms in greater detail. An instructor could use pretest data on the LOCI to identify any misconceptions commonly held by students and then tailor his or her teaching appropriately. Posttest data could then be used with pretest data to observe any learning gains made by students. Raw scores and learning gains are two ways to interpret LOCI scores.
Another method of evaluating scores on the LOCI is to observe the breakdown of scores by learning objective. The same score, a six out of 12, for example, would mean different things if the six correct items were grouped in one or two learning objectives versus being spread evenly across all learning objectives. Using this information, an instructor has the ability to determine that students do not understand an entire learning objective or have specific areas of confusion within a learning objective. This utility informs teaching in a practical way, allowing instructors to pinpoint students’ needs.
The LOCI covers a broad range of difficulty and has the ability to discriminate between high- and low-performing students, and the data obtained are predicted to be very reliable. From the perspective of item response theory, this manuscript demonstrates the utility of the LOCI as a tool for both educators and researchers in assessing student understanding of lac operon function.
However, overall, students performed poorly on the LOCI following traditional instruction. The average score for all participants was 5.65 out of 12 (or 47.1%), indicating that, in general, these students struggled with this concept. This lends support to the need for additional or improved instructional techniques to help students gain more understanding of this complicated regulatory system. The misconceptions that were identified can provide instructors with insight into student thinking and better inform their approaches. Once common misconceptions are inventoried and identified, models of instruction based on conceptual change can be utilized (Bybee, 2002). For example, knowing that students have confusion over the location to which the repressor protein binds, instructors can construct appropriate hands-on or visual activities that cause students to confront this misconception. Our own work has demonstrated this to be true (Stefanski, 2015)
Active-learning strategies have been shown to be effective and are included among promoted strategies of instruction (AAAS, 2011; Tanner, 2013). Despite reports like Vision and Change calling for biology instructors to adopt more active-learning techniques into their courses, widespread change has yet to be achieved (Tagg, 2012; National Research Council, 2013). Biology faculty members have indicated, among other factors, that they feel ill-equipped to enact these changes due to a lack of sufficient training (Brownell and Tanner, 2012). By providing ready-to-use assessments like the LOCI, we can better equip instructors to generate, assess, and utilize evidence-based active-learning resources that target difficult concepts. In her 2013 follow-up to Vision and Change, D’Avanzo calls for educators to take advantage of the existing body of CIs to provide evidence of program efficacy (D’Avanzo, 2013). She suggests that learning gains calculated by using CIs be tied to one or more of the five conceptual areas laid out in Vision and Change and that these data be presented to faculty members to influence their development of course design (D’Avanzo, 2013). In summary, this study addresses the barriers to change noted above by providing the means of gathering evidence regarding the instruction and conceptual understanding of the valuable and far-reaching concept of gene regulation.



