Implementation of a Collaborative Series of Classroom-Based Undergraduate Research Experiences Spanning Chemical Biology, Biochemistry, and Neurobiology
Abstract
Classroom undergraduate research experiences (CUREs) provide students access to the measurable benefits of undergraduate research experiences (UREs). Herein, we describe the implementation and assessment of a novel model for cohesive CUREs focused on central research themes involving faculty research collaboration across departments. Specifically, we implemented three collaborative CUREs spanning chemical biology, biochemistry, and neurobiology that incorporated faculty members’ research interests and revolved around the central theme of visualizing biological processes like Mycobacterium tuberculosis enzyme activity and neural signaling using fluorescent molecules. Each CURE laboratory involved multiple experimental phases and culminated in novel, open-ended, and reiterative student-driven research projects. Course assessments showed CURE participation increased students’ experimental design skills, attitudes and confidence about research, perceived understanding of the scientific process, and interest in science, technology, engineering, and mathematics disciplines. More than 75% of CURE students also engaged in independent scientific research projects, and faculty CURE contributors saw substantial increases in research productivity, including increased undergraduate student involvement and academic outputs. Our collaborative CUREs demonstrate the advantages of multicourse CUREs for achieving increased faculty research productivity and traditional CURE-associated student learning and attitude gains. Our collaborative CURE design represents a novel CURE model for ongoing laboratory reform that benefits both faculty and students.
INTRODUCTION
Recent high-profile presidential and influential national scientific panels have appealed for widespread incorporation of novel research experiences into undergraduate laboratory curricula and the development of interdisciplinary, collaborative undergraduate educational opportunities (National Research Council [NRC], 2003; President’s Council of Advisors on Science and Technology [PCAST], 2012; American Academy of Arts and Sciences, 2013). One tractable and flexible model for broadly introducing collaborative research experiences into the science curriculum is classroom undergraduate research experiences (CUREs; Auchincloss et al., 2014; Corwin et al., 2015). CUREs provide student populations access to the measurable benefits of undergraduate research experiences (UREs) while balancing faculty, student, and institutional roadblocks to involvement in UREs (NRC, 2003; PCAST, 2010, 2012; American Association for the Advancement of Science, 2011; Auchincloss et al., 2014; Corwin et al., 2015; Linn et al., 2015). CUREs also have multiple confirmed student outcomes, ranging from direct, course-related outcomes (increased content knowledge, increased analytical skills, and increased technical skills) to personal and career-related achievements (increased self-efficacy, external validation, persistence in science, and career clarification) (Beck et al., 2014; Corwin et al., 2015; Linn et al., 2015). Importantly, these benefits are particularly pronounced in participants from underrepresented groups (Russell et al., 2007; Carter et al., 2009; Bangera and Brownell, 2014; Linn et al., 2015).
A wide variety of CUREs have been developed and assessed; these range from large multi-institutional CUREs to single-section, single-institution CUREs (Hatfull et al., 2006; Wu, 2013; Buonaccorsi et al., 2014; Jordan et al., 2014; Lopatto et al., 2014; Brownell et al., 2015; Corwin et al., 2015; Russell et al., 2015; Shapiro et al., 2015). The target demographics for CUREs also vary across the undergraduate spectrum from introductory major and nonmajors courses to upper-level multisemester courses (Nadelson et al., 2010; Brownell et al., 2015; Rowe et al., 2015). High-profile CUREs, including the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) and the Genomics Education Partnership (GEP), have been highly successful at linking together introductory laboratory experiments across multiple institutions with common experimental procedures and producing significant scientific and student benefit outcomes (Shaffer et al., 2010; Jordan et al., 2014; Lopatto et al., 2014).
Despite the evidence supporting the success of CUREs in promoting student learning and engagement, challenges to implementation still exist. Chief among these with regard to CURE laboratories are the time and energy required for lab design and implementation, which must be balanced with the various other responsibilities of faculty who have both research and teaching obligations (Darden, 2003; Kloser et al., 2011; Lopatto et al., 2014). Rather than designing a CURE on an entirely new project with which a faculty member has little prior experience, incorporating a project that builds on current work being done in a faculty member’s own research laboratory has the potential to facilitate research progress for the faculty member, while also maximizing student learning in the CURE (Darden, 2003; Kloser et al., 2011; Brownell and Kloser, 2015). However, despite this call, few reports of courses designed specifically around faculty research interests have been reported, and those courses that have been reported focused entirely on data collection, were multi-institutional network CUREs with unique challenges from independent CUREs, or have yet to be assessed for their dual effects on both student learning and faculty research productivity (Nadelson et al., 2010; Kloser et al., 2011; Ditty et al., 2013; Miller et al., 2013; Lopatto et al., 2014; Shortlidge et al., 2016).
With the confirmed benefits of CUREs to students and faculty (Wu, 2013; Auchincloss et al., 2014; Harvey et al., 2014; Corwin et al., 2015), a limited number of institutions have begun to develop CURE programs that span multiple courses or even an entire curriculum (Knutson et al., 2010a,b; Russell et al., 2015; Shapiro et al., 2015; Sanders et al., 2016). These larger-scale CUREs provide tractable models of modern, collaborative science and move toward the complex, interdisciplinary nature of scientific investigation. For example, an integrated CURE-based curriculum was recently described that jointly studied insect biodiversity across a sophomore-level cell biology course and a junior-level ecology course, with sharing of resources, results, and learning outcomes to produce significant student benefits and novel research products (Russell et al., 2015). The competency-based research laboratory curriculum (CRLC) at the University of California, Los Angeles (UCLA), also provided divergent CUREs or apprentice-based research experiences (AREs) to every student across the curriculum (Shapiro et al., 2015; Sanders et al., 2016). These separate CURE and ARE tracks produced similar student outcomes for each curriculum, with the CURE track proving particularly effective in reducing the achievement gap between high- and low-performing students (Shapiro et al., 2015). The four upper-division CUREs in CRLC spanned the biology curriculum, from courses in microbiology to plant–microbe ecology, and shared common student learning outcomes, but had divergent research goals (Sanders et al., 2016). At institutions with significant research and teaching requirements, further development of such interdisciplinary and collaborative CUREs could assist faculty in simultaneously engaging in innovative teaching and research in a manner that minimizes time and resource barriers while maximizing faculty productivity and student learning benefits.
Herein, we describe the implementation of a novel set of three upper-division CURE laboratories that incorporate collaborative faculty research across multiple departments spanning the scientific disciplines of chemical biology, biochemistry, and neurobiology. These courses engaged students in novel scientific research questions involving the development and use of fluorescence detection tools to investigate various aspects of Mycobacterium tuberculosis infection or neural signaling (current research topics investigated by the faculty running these courses) and produced significant publishable and presentable research outcomes. Building on previous cross-curricular CURE models, our multi-CURE design is the first CURE model focused on the collaborative integration of faculty research projects across departmental and disciplinary boundaries into a series of CUREs.
We set the following five scientific and pedagogical goals for successful implementation of our collaborative CURE curriculum: 1) generate novel scientific data related to bacterial serine hydrolases and neuronal signaling; 2) develop students’ experimental design and data analysis skills; 3) promote positive student attitudes about science and perceptions of learning gains; 4) promote student retention in science, technology, engineering, and mathematics (STEM) disciplines; and 5) promote faculty research productivity. On the basis of faculty, student, and scientific outcomes, we think that this collaborative CURE model, which incorporates interdisciplinary faculty research interests, provides a general outline for bridging faculty teaching and research expectations while maintaining strong student learning outcomes.
METHODS
Context
The collaborative series of three CUREs included a new laboratory course in chemical biology (CHEMBIO), a revised laboratory course in biochemistry (BIOCHEM), and the laboratory component of a new lecture plus laboratory course in neurobiology (NEURO) at Butler University, a primarily undergraduate institution in Indianapolis, Indiana. All three CURES were elective courses within their respective majors; NEURO also was a required course in a recently developed neuroscience minor. Thus, students largely self-selected into these courses. Each course ran three times between the Fall of 2012 and the Spring of 2015 and was tailored toward upper-level science students and small class sizes (eight to 16 students).
Participants
Participating in each course were junior- and senior-level biology, chemistry, and psychology students (Supplemental Table S1). Prerequisites included two semesters of organic chemistry (lecture plus lab) for CHEMBIO, one semester of biochemistry lecture and analytical chemistry (lecture plus lab) for BIOCHEM, and one semester of lecture plus lab courses in genetics and cell and molecular biology for NEURO (Figure 1C). Beyond these prerequisites, students self-selected into these courses and were able, but not required, to take all three courses. No CURE was a prerequisite for any other course. Fifteen of the 46 total students (34%) enrolled in either CHEMBIO or BIOCHEM took both courses; however, there was no overlap between the students in either of these courses with the students who took NEURO. No students were enrolled simultaneously in more than one CURE. Course instructors were full-time tenure-track faculty in the departments of chemistry (CHEMBIO, BIOCHEM; G.C.H. and R.J.J.) and biological sciences (NEURO; J.R.K.). Each of the two chemistry faculty members (G.C.H. and R.J.J.) taught CHEMBIO and BIOCHEM at least once. The same faculty member in biological sciences taught all three iterations of NEURO (J.R.K.). At the time the courses were taught, one faculty member in chemistry was tenured (G.C.H.), while the other two faculty members were untenured (J.R.K. and R.J.J.). All had active research programs related to the areas being investigated in the CUREs; research is a requirement for tenure at Butler.
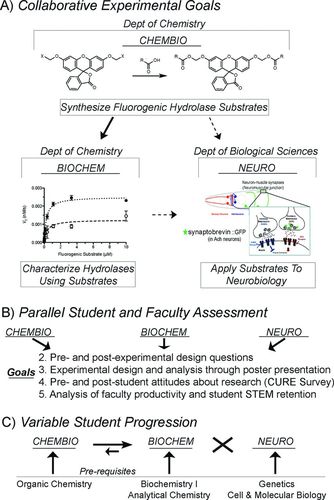
FIGURE 1. Overview of experimental theme, faculty and student assessment strategy, and student progression through CHEMBIO, BIOCHEM, and NEURO. (A) Collaborative experimental goals: CHEMBIO and BIOCHEM (offered through the Department of Chemistry) and NEURO (offered through the Department of Biological Sciences) laboratories revolved around the central theme of visualizing biological processes like M. tuberculosis enzyme activity and neural signaling using fluorescent molecules. In this collaborative design, CHEMBIO synthesized bis-acyloxymethyl ether derivatives of fluorescein and BIOCHEM and NEURO tested different biological applications of these fluorogenic compounds. Although the fluorophores were impermeable to the C. elegans cuticle and were not fully integrated into NEURO (as reflected by the dashed arrow), NEURO used traditional fluorescence methods in combination with RNAi methods to test the role of specific enzymes in controlling neural signaling. (B) Parallel faculty and student assessments: As each CURE followed a similar course design (see Table 1), parallel assessments were used to assess student learning of specific course content and common outcomes for both faculty and students. Goals are numbered to match our five goals. Goal 1 was to “generate novel data related to bacterial serine hydrolases and neuronal signaling” and is reflected by the schematic in A. (C) Variable student progression: While the experimental goals were collaborative, each of the three CUREs was a stand-alone course designed to provide similar experimental training–related outcomes for students. Thus, while students were able to take any or all of the three CUREs for which they met the prerequisites (shown in the lower boxes) and some students progressed from CHEMBIO to BIOCHEM, only a few students who took BIOCHEM later took CHEMBIO, and no students took NEURO and CHEMBIO or BIOCHEM. The width of the arrows represents the relative student movement between courses, with the “X” indicating the lack of cross-movement between NEURO and the other two courses.
Curriculum Design
Our three CUREs met the accepted definition of CUREs and were designed to have collaborative experimental goals and parallel course assessments but to allow for variable student progression (Figures 1 and 2; Table 1; Auchincloss et al., 2014). Each CURE transitioned students from initial experiments based on questions and procedures provided by the instructors to independent projects in which students selected the questions and designed the experiments. In these laboratories, students investigated globally relevant scientific research questions that developed interdisciplinary, collaborative faculty research interests. The collaborative experimental goals included the development and use of a class of biological imaging agents known as fluorogenic hydrolase substrates to characterize the activity of a class of enzymes known as serine hydrolases from M. tuberculosis (TB) and to investigate the molecular control of neuronal signaling in Caenorhabditis elegans (Lavis and Raines, 2008; Long and Cravatt, 2011; Dedieu et al., 2013). Specifically, students synthesized the fluorogenic substrates in CHEMBIO, used these imaging agents to catalogue the substrate specificity of serine hydrolases from TB in BIOCHEM (Johnson et al., 2014a,b; Lukowski et al., 2014), and initially tried to combine the products of the two labs to screen for new components of neuronal signaling in C. elegans in NEURO (Figure 1A). Although the initial goal of having NEURO students use the newly characterized hydrolase–fluorophore substrate pairs as imaging tools to investigate neuronal signaling was limited by the permeability of the fluorophores to the C. elegans nervous system (Tian et al., 2012), NEURO maintained similar methodologies and overlapping pedagogical design and goals. CHEMBIO and BIOCHEM maintained the common scientific theme and shared research products (Figure 1A).
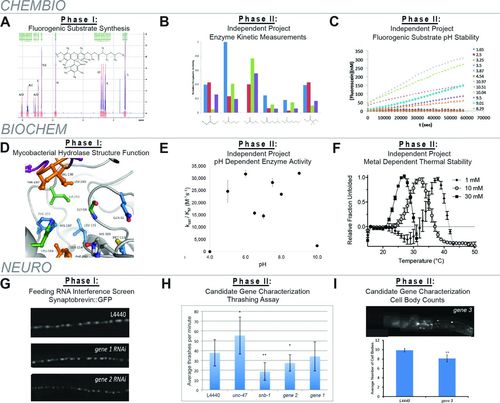
FIGURE 2. Sample student data from each CURE. (A) CHEMBIO sample synthetic data (phase I). Students completed a three-step synthesis of bis-acyloxymethyl ethers from fluorescein with an SN2 reaction (Figure 1A). Shown is the 1H-NMR spectrum for the ethylthiopropanyl derivative. Students also characterized their products by high-resolution MS and 1H-NMR/13C-NMR/IR spectroscopy. (B and C) CHEMBIO independent project results (phase 2). (B) Overall catalytic efficiency of six student-synthesized fluorogenic substrates against four different serine hydrolases (blue: pig liver esterase control; red: Rv0045c from M. tuberculosis; green: LipW from M. marinum; purple: FTT0258 from Francisella tularensis). (C) Chemical stability of student-synthesized fluorogenic substrate measured over 16 h from pH 1.65 to pH 11. (D) BIOCHEM combined student data (phase 1). A three-dimensional depiction of the binding pocket of the Rv0045c serine hydrolase from M. tuberculosis (PDB: 3P2M). Each of the residues shown in the ball-and stick-representation was individually substituted to alanine by different BIOCHEM students, and kinetic data from BIOCHEM students’ purified proteins were combined to create a coherent story for the overall course (Lukowski et al., 2014). (E and F) BIOCHEM phase 2 independent project results. (E) Overall catalytic efficiency of student-purified LipN from M. ulcerans measured across a range of pH values using a fluorogenic hydrolase substrate. (F) Decrease in the thermal stability of Rv0045c from M. tuberculosis dependent on increased concentrations of cobalt. (G) NEURO sample RNAi screen data (phase 1). Knockdown of ubiquitin ligase genes (genes 1 and 2) alters synaptobrevin::GFP localization and abundance in cholinergic motor neurons of young adult C. elegans worms (L4440 = negative control). (H and I) NEURO independent project results (phase 2). (H) Results of a thrashing assay performed on animals treated with RNAi targeting each of the two genes in G, unc-47, which is involved in inhibitory GABA release, and snb-1, which regulates synaptic vesicle release in all neurons (n ≥ 10 animals per treatment; Student’s t test vs. L4440 controls: *, p < 0.01; **, p < 0.00001). (I) Results of cholinergic motor neuron cell body counts performed on animals treated with RNAi targeting gene 3, which exhibited decreased density of synaptobrevin::GFP-labeled cholinergic synapses in the phase 1 RNAi screen (unpublished data). Images show representative ventral nerve cords and cell bodies; graphs depict average neuron counts (control L4440 [n = 8] or gene 3 [n = 16]; Student’s t test vs. L4440 controls: **, p < 0.00001). Error bars show ± SD of the mean for each measurement.
| CHEMBIO | BIOCHEM | NEURO |
|---|---|---|
| Phase 1 | ||
| Synthetic design (1–2) | Molecular biology (1–4) | C. elegans training/gene selection (1–2) |
| Choose synthetic target compounds and starting materials | Design mutations; design primers; molecular model | Practice C. elegans picking and maintenance; select RNAi genes |
| Synthesis (3–5) | Protein purification (5–6) | RNAi protocol design (3–5) |
| Optimize reaction and chromatography purification conditions | Choose buffers; choose induction conditions | Plan/optimize RNAi treatment protocol; microscopy training |
| Chemical characterization (7–9) | Protein characterization (7–9) | RNAi microscopy screen (6–9) |
| Choose NMR/IR conditions; collect spectra; prepare MS samples | Choose kinetic substrate; choose temperatures | Collect microscopy data, analyze images |
| Phase 2 | ||
| Independent projects (10–13) | Independent projects (10–13) | Independent projects (10–13) |
| Develop hypothesis; design experiments; prepare reagents; analyze results | Develop hypothesis; design experiments; prepare reagents; analyze results | Develop hypothesis; design behavorial/imaging/RNAi assay; analyze results |
| Poster presentation (14) | Poster presentation (14) | Poster presentation (14) |
While students in each CURE worked independently to tackle aspects of these interdisciplinary scientific questions, cross-communication occurred at interdepartmental poster sessions. Student and faculty outcomes from each CURE were analyzed using parallel assessment rubrics to reinforce the expected student, faculty, and scientific benefits of the CUREs (Figure 1B). Unlike other collaborative course designs, the curricular structure of these three collaborative, interdisciplinary CUREs was not constructed to shuttle students across the three courses but rather to facilitate faculty collaboration while providing flexibility in student CURE involvement to increase student participation (Figure 1C). Together, these collaborative CUREs and the overall curricular design provide a new CURE model based on scientific collaboration among interdisciplinary faculty with curricular flexibility and universal student benefits (Figure 1).
Course Descriptions and Implementation
CHEMBIO Course.
CHEMBIO was a newly developed, stand-alone upper-level chemistry laboratory course. CHEMBIO met once per week for 4 hours and was populated by an average of eight upper-level chemistry majors per semester. The experimental goals of CHEMBIO were to synthesize a series of related fluorogenic compounds and to then subject those compounds to chemical and/or biological characterization. CHEMBIO served as the starting point for the larger experimental goals of using the fluorogenic compound library to characterize mycobacterial serine hydrolases (BIOCHEM) and to identify novel ubiquitin ligases in C. elegans (NEURO). The synthetic target compounds for CHEMBIO were bis-acyloxymethyl ether derivatives of fluorescein, previously shown to be highly stable enzyme probes (Figure 1A; Lavis et al., 2011; Tian et al., 2012; Lukowski et al., 2014). Introduction of a serine hydrolase to these protected fluorogenic substrates catalyzes their activation, with the relative efficiency of enzyme-catalyzed hydrolysis dependent upon the specific recognition of the acyl-chain structure of these fluorogenic esters by the serine hydrolase (Lavis et al., 2011; Hedge et al., 2012; Tian et al., 2012). Thus, a series of bis-acyloxymethyl ether derivatives of fluorescein can serve as a tool for characterizing the substrate specificity of mycobacterial serine hydrolases (Hedge et al., 2012; Ellis et al., 2013; Filippova et al., 2013; Johnson et al., 2014a; Lukowski et al., 2014) and for developing novel neural-signaling sensors (Tian et al., 2012).
To construct this library of protected fluorogenic hydrolase substrates, students in CHEMBIO for phase 1 each pursued two novel bis-acyloxymethyl ether derivatives of fluorescein. A common synthetic methodology was pursued by all students: the SN2 displacement of chloride leaving groups on a common precursor compound (Figure 1A). CHEMBIO students thus collectively used a combinatorial synthetic approach to expand an ever-larger library of bis-acyloxymethyl ether derivatives of fluorescein. Students designed their own target compounds using various strategies, ranging from incremental structural variations of existing compounds in the library to computer-assisted molecular modeling (Table 1, weeks 1–2). Students then chose their experimental conditions for the synthetic reactions (reagent ratios; reaction time; solvent) and conditions for monitoring reaction progress via TLC (solvent mixture; visualization). Products were isolated from reaction mixtures by liquid chromatography under conditions chosen by the students based on TLC data (Table 1, weeks 3–5). Purified products were then characterized via proton nuclear magnetic resonance (1H-NMR; sample student data in Figure 2A), 13C-NMR, mass spectrometry (MS), and infrared (IR) spectroscopy (Table 1, weeks 7–9).
After confirmation of the correct synthesis of the desired fluorogenic substrates, CHEMBIO then transitioned in phase 2 to student-driven independent research projects using the synthetic products (Table 1, weeks 10–13). Independent projects designed by the students could be grouped into four general categories: 1) repeating the synthetic procedure for failed synthesis or refinement of library compounds, 2) investigation of the physicochemical stability of their fluorogenic product compounds under various conditions (temperature, pH; sample student data in Figure 2C), 3) measurement of the cellular stability of these fluorogenic compounds following uptake into mammalian or bacterial cells, 4) and characterization of the steady-state kinetics for fluorogenic substrate hydrolysis by various hydrolase panels (sample student data in Figure 2B).
Final results of the independent projects were shared at a semiannual end-of-semester departmental poster session of students and faculty from the biological sciences and chemistry departments (Table 1, week 14). The poster presentations provided an opportunity for students from the three courses to collaboratively discuss their research results and to follow the progression of the projects throughout the years.
BIOCHEM Course.
Building on the work of students in CHEMBIO, BIOCHEM students used the highest-activity fluorogenic hydrolase substrate from CHEMBIO to precisely characterize the active site of three different serine hydrolases. These three serine hydrolases were selected due to their interesting chemical biology properties and the critical health impact of TB (Dedieu et al., 2013; World Health Organization, 2015). BIOCHEM was a stand-alone upper-level chemistry laboratory with eight to 12 junior/senior chemistry or biology majors per semester. The laboratory met once per week for 4 hours and also fulfilled university requirements for a writing-intensive course. The experimental design for BIOCHEM paralleled similar semester-long experiments at other institutions relating the structure and function of a protein using site-directed mutagenesis, but fused this overall course design with the larger scientific goal of characterizing important mycobacterial serine hydrolases (Bailey, 2009; Knutson et al., 2010a; Kreiling et al., 2011).
For the 3 years of the course assessed here, the structure and function of three different mycobacterial hydrolases were characterized, with each year’s class undertaking the analysis of a different serine hydrolase. A significant number of serine hydrolases have been identified as important for lipid metabolism by TB, but only a few have been purified and/or have had their enzymatic activity verified (Singh et al., 2010; Dedieu et al., 2013). To begin the semester, each BIOCHEM student was assigned two specific mutations within the active site or binding pocket of the chosen mycobacterial serine hydrolase and was provided with mutagenic primers to convert each amino acid to alanine (Table 1, weeks 1–4). Approximately 10–14 total mutations for the course were targeted per semester so that, with two mutations per eight students, a significant degree of redundancy was included in the course design. During phase 1, BIOCHEM students then combined the structural information about their assigned residues with the biochemical data about their resulting proteins to construct hypotheses about the roles of their amino acids in the structure and function of the mycobacterial serine hydrolase (Table 1; Figure 2D; Lukowski et al., 2014). The pinnacle of phase 1 was the biochemical characterization of the final purified hydrolase variants using thermal stability and enzymatic catalysis measurements with the highest-activity fluorogenic substrate identified in each year of CHEMBIO (Table 1, weeks 5–9). Detailed experimental procedures for these last two laboratory experiments and example student data were published and together illustrate the generalizability of these experimental procedures (Johnson et al., 2014a,b).
During phase 2, which encompassed the final 4 weeks of the semester, students worked in groups of twos and threes to design their own independent projects related to the mycobacterial serine hydrolase (Table 1, weeks 10–13). To plan their independent projects, students wrote short (one to two pages) scientific proposals or outlines describing their hypothesis, the significance of their project, and a proposed methodology. BIOCHEM faculty mentors then assisted students in finalizing their experimental designs based on individual meetings. BIOCHEM students designed independent projects ranging from direct extensions of the semester’s laboratory experiments, including determining the effect of various metals, solvents, temperatures, inhibitors, or additional fluorogenic substrates from CHEMBIO on the catalytic activity of the mycobacterial serine hydrolase, to more divergent projects, including cloning homologous serine hydrolases, multiple sequence alignments, crystallography trials, and synthesis of refined fluorogenic substrates (Figure 2, E and F; Johnson et al., 2014a,b). At the end of the semester, students presented the results from their independent projects to fellow students and faculty at the joint biological sciences and chemistry departmental poster session (Table 1, week 14).
NEURO Course.
Continuing the collaborative work of CHEMBIO and BIOCHEM, the original research goal of NEURO was to study neural signaling using the fluorogenic substrates from CHEMBIO. For NEURO, the CURE comprised the laboratory portion of a new upper-division course in cellular and molecular neuroscience. The more traditional lecture/discussion component of the course met twice a week for 75 minutes, while the laboratory met for one 3-hour session per week. NEURO was taken by five to 15 junior and senior biology and psychology majors per semester. NEURO is distinct from the CHEMBIO and BIOCHEM courses due to the added lecture/discussion component, which is intertwined with the lab in terms of the focus on experimental design, data analysis, and techniques associated with cellular and molecular neuroscience. The overarching research goal of NEURO was to identify and begin to characterize novel ubiquitin family enzymes that regulate neural signaling, using the model roundworm C. elegans (Figure 1A). Ubiquitin ligases and proteases (DUBs) are required for synapse development, maintenance, and function, and for neural development in C. elegans (Bingol and Sheng, 2011; Kowalski and Juo, 2012). Many ubiquitin ligases are expressed in the nervous system; however, the function, regulation, and substrates of most of these enzymes in neurons and at synapses have not yet been investigated.
To identify novel ubiquitin ligases with roles in controlling synaptic signaling, NEURO students worked in groups of three or four in phase 1 to perform RNA interference (RNAi)-based screens of transgenic worms expressing the fluorescently tagged synaptic vesicle protein synaptobrevin::green fluorescent protein (synaptobrevin::GFP) in cholinergic motor neurons (Table 1; Zhen and Jin, 1999; Sieburth et al., 2005). These labeled synaptic vesicles localize in clusters at presynaptic sites adjacent to postsynaptic muscle cells at neuromuscular junctions (Figure 1A). The initial plan for NEURO was to have students screen for effects of ubiquitin ligase knockdown on the abundance of a serine hydrolase–tagged receptor that would activate exogenously delivered masked fluorogenic substrates from CHEMBIO in a cell-specific manner; however, due to permeability issues with the worm cuticle, the approach was switched to a traditional GFP-based imaging system. Before performing the RNAi screen, each group first researched and selected 10 ubiquitin ligase genes to test from a list of 25 provided, while also reviewing sterile technique and optimizing the RNAi treatment procedure (Table 1, weeks 1–4). Groups then spent several weeks performing visual screens of synaptobrevin::GFP-expressing worms in which they had individually inhibited the expression of each of their chosen ubiquitin ligase genes via RNAi (Table 1, weeks 5–9, and Figure 2G). Following the screening period, individual students each wrote and revised a scientific manuscript describing their phase 1 screen, and each group selected one to three candidate genes for further characterization during phase 2 (Table 1, weeks 10–13).
In phase 2, the student groups designed independent projects with which to further investigate enzymes selected based on the results of their phase 1 RNAi screens. These projects included reiterative quantitative imaging, enhancer/suppressor RNAi screens, behavioral assays to assess synapse function, and additional imaging analyses to test for effects of their candidate ubiquitin enzyme(s) on another synapse (Figure 2, H and I). Before the start of phase 2, each group submitted a brief research proposal outlining a hypothesis, experimental methods, expected results and anticipated problems; the proposal was revised and approved before experimentation began. As with CHEMBIO and BIOCHEM, the project culminated with group poster presentations at the joint biological sciences and chemistry poster session (Table 1, week 14).
Syllabi and a detailed week-by-week description of activities for each course, can be found in the Supplemental Material. Additional protocols and course assignments are available upon request.
Assessment Data Collection and Analysis
Human Subjects Protocol.
The Butler University Institutional Review Board approved the research described in this work as an exempt protocol (May 2011). The number of students in each course who participated in each assessment is given in legends for the figures in which those data are described.
Experimental Design Assessment.
To measure the effects of student participation on the development of students’ experimental design skills, students provided written answers to a pair of course-specific short-essay questions on the first (precourse) and last (postcourse) day of class (see the Supplemental Material). These questions were designed specifically to test students’ ability to design and interpret data from experiments similar to those they had encountered in their semester-long research projects and tested both specific methodological understanding relevant to their projects and fundamental aspects of experimental design, such as the use of positive and negative controls. We chose to use these questions, written by the instructor of each course, rather than pre-established assessment tools, due to the complex nature of the science investigated in each course and our desire to teach discipline-specific molecular techniques in addition to general experimental design skills.
All student responses to the pre- and postcourse experimental design questions were collected anonymously and were independently scored by each of the three course instructors using a four-point scale based on pretested rubrics (see the Supplemental Material) that were examined by another faculty member in the chemistry department who was uninvolved in the study. The three scores per student were then averaged to obtain a single score for each student. Instructors were not blinded to pre- versus postcourse question sets. Student scores across all three sections of a given course were then averaged for both the precourse questions and the postcourse questions, and the means of pre- and postcourse scores were statistically compared as described in Statistical Analyses. Because student responses were not coded, pre- and postcourse responses for a single student could not be compared. The anonymity with which student responses were collected also prevented tracking of students who took more than one of the CUREs as opposed to those who took only one course.
Scientific Poster Assessment.
Students in all courses worked in groups of two to four students to prepare posters describing their independent projects, which they presented at a public poster session at the end of the semester. These posters were evaluated using a common, pretested rubric (see the Supplemental Material) examined by an outside evaluator in the chemistry department. Each component of the poster was rated on a scale from 0 (component not present) to 3 (component fully meets expectations), in which all information is included and described clearly, with appropriate connections and depth. Poster components included 1) basic poster formatting; 2) articulation of project significance; 3) clear statement of hypothesis or project goal; 4) experimental methods that are sound and well-controlled; 5) accurate, complete data presentation; 6) conclusions given in light of original goals/hypothesis; and 7) appropriate future modifications and extensions of the project. All posters were evaluated by all three instructors, and the average poster score in each of the seven areas was determined for each course over all three iterations. The instructors graded all posters, as they were the relevant content experts in the disciplines covered in the CUREs. Individual instructor/course bias was minimized by having all three instructors grade all posters from all three courses using a standardized rubric. While all students were required to participate in poster preparation and presentation, only the aggregate score for each poster group was collected, as the goal of the assessment was to quantitate the scientific communication, experimental design, and data analysis of the class as a whole.
CURE Assessment of Student Attitudes and Perceived Learning Gains.
Students participated in the online CURE survey (www.grinnell.edu/academics/areas/psychology/assessments/cure-survey) both pre- and postcourse; this survey measures student attitudes about science and student perceptions of their own learning gains (Lopatto, 2004, 2007; Lopatto et al., 2014). We acknowledge the recent questions regarding the validity and reliability of the CURE survey instrument in accurately measuring student attitudes and perceptions across a variety of CUREs and contexts and the potential effects of social bias and differing levels of feedback on student CURE responses (Auchincloss et al., 2014). However, the CURE survey method was chosen because this method allows for comparison of the attitudes and perceived learning gains of the students in each of our three CUREs with those of students in other CUREs and with those of students who participated in summer undergraduate research experiences (SUREs) across the country. Data analysis was performed by the CURE survey group at Grinnell College and included the SDs for the aggregate CURE and SURE data across institutions and courses.
Student Research Retention.
Student retention in research was measured by determining 1) the number of CURE students who participated in independent undergraduate research projects either at Butler or SUREs or internships at other institutions and 2) the number who completed honors theses. Student retention in STEM fields was also assessed by determining the number of students attending or planning to attend graduate school or professional school in STEM-related fields or continuing in a STEM-related job postgraduation.
Faculty Research Productivity Assessment.
Faculty research output was measured by calculating 1) the average number of research students mentored per semester by all three faculty members, 2) the total number of peer-reviewed research or pedagogical publications (in print, accepted, or under revision), and 3) the total numbers of research or pedagogical presentations (posters or oral presentations) made by each faculty member and/or his or her students at regional or national scientific conferences. Precourse faculty output counts were made from Fall 2009 through Summer 2012; postcourse counts were made from Summer 2013 through Spring 2016 to allow a lag period for completion of one iteration of each course before postcourse faculty output measurements. Numbers of presentations and publications involving undergraduate coauthors are indicated later in this article in Table 3, as are the number of faculty-student coauthored publications directly resulting from the CUREs and related research projects.
Statistical Analyses
The SPSS statistical analysis program was used to perform independent variables t tests for statistical significance testing and Cronbach’s alpha interrater reliability testing on pre- and postcourse experimental design questions and Cronbach’s alpha testing on poster scores. The JMP statistical analysis program was used to perform a nonparametric Kruskal-Wallis (Wilcoxon) test with a one-way chi-square approximation on the mean research student numbers per semester for all three faculty members combined, as the combined precourse data were not normally distributed. JMP software was also used to perform unpaired two-tailed t tests on the research student data for individual faculty members following tests for equality of variances. The alpha level for statistical significance of all tests was 0.05.
RESULTS AND DISCUSSION
Integration of research experiences into classroom laboratories (CUREs) has been a major focus of recent revisions to undergraduate science curricula, given the documented benefits to student engagement, confidence, critical-thinking skills, and retention in science (Auchincloss et al., 2014; Bangera and Brownell, 2014; Brownell and Kloser, 2015; Brownell et al., 2015; Corwin et al., 2015). Incorporation of faculty members’ research projects into CUREs can also decrease implementation efforts and maximize research and teaching benefits to faculty while promoting student learning and engagement outcomes (Darden, 2003; Nadelson et al., 2010; Kloser et al., 2011; Ditty et al., 2013; Miller et al., 2013; Russell et al., 2015). Considering these measurable benefits of CUREs to faculty and students and the lack of CUREs in the molecular life science curriculum at Butler University, we implemented a series of three collaborative CUREs focused on big-picture topics of societal and scientific importance and with direct connections to ongoing faculty research projects (Figure 1). These CUREs involved collaborative research and teaching approaches across three upper-division laboratory courses in the biological sciences and chemistry departments, and each engaged students in independent research projects requiring significant student input into the questions, experimental designs, data analysis, and presentation of the findings of these projects. For this curricular design, we developed and assessed five specific goals based on successful implementation of these collaborative CUREs (Figure 1).
Goal 1: Generate Novel Data Related to Bacterial Serine Hydrolases and Neuronal Signaling
CHEMBIO.
In CHEMBIO, students designed, synthesized, and chemically characterized the properties of their fluorogenic substrates (Figures 1 and 2). Building on their skills from organic chemistry, students applied synthetic methodology to a novel chemical problem and obtained experience with NMR, different MS techniques, and flash chromatography (Figure 2A). At the beginning of the semester, faculty and students worked collaboratively to design a novel series of fluorogenic substrates using various strategies, ranging from incremental structural variations of existing compounds in the library to computer-assisted molecular modeling. Through 3 years of CHEMBIO, the published library of fluorogenic substrates was expanded from ∼20 to more than 50 substrates, opening up the scientific questions that can be addressed with this library. Each of these substrates has detailed chemical characterization, including 13C-NMR, 1H-NMR, and high-resolution MS, and was synthesized in sufficient quantities for all future applications. Because these substrates served as the starting point for further investigations for mycobacterial hydrolases from BIOCHEM and neural signaling in NEURO, scientific outcomes are still forthcoming based on the application of the CHEMBIO substrates, but two publications using the fluorogenic substrates from CHEMBIO are currently in preparation.
Phase 2 independent projects from CHEMBIO have proven fruitful initiation points for future investigations, including understanding the relatively large shifts in the stability of the fluorogenic substrates based on changes in pH and temperature and developing comprehensive structure-activity relationships for mycobacterial serine hydrolases (Figure 2, B and C). These independent projects and refinement of final synthetic products have been the subject of two senior theses and multiple national scientific presentations from students continuing their research from CHEMBIO into an independent research project (see goal 5). Additionally, two students from CHEMBIO continued into independent research in the summer after they graduated from Butler, and one of these students continued into graduate school in biological chemistry after this summer research experience.
BIOCHEM.
BIOCHEM laboratory followed a classic procedure for creating site-directed protein variants and then studying the effect of these amino acid substitutions on the structure and function of an enzyme (Knutson et al., 2010a,b). The novelty of BIOCHEM is the focus of the structure–function studies on biomedically relevant serine hydrolases from M. tuberculosis and the direct connection with the fluorogenic substrates from CHEMBIO. As an example of the novel scientific results from BIOCHEM, students in the first iteration of BIOCHEM constructed alanine-scanning variants of the binding pocket and active site loop of the mycobacterial hydrolase Rv0045c (Figure 2A). From this initial alanine screen, two key residues (Gly-90 and His-187) were identified that increased the catalytic activity of Rv0045c toward varied fluorogenic substrates. The following cycle of BIOCHEM then built on this initial screen and performed saturation mutagenesis of these key residues, finding an evolutionarily substituted tyrosine that increased the catalytic activity of Rv0045c greater than 20-fold. Combination of these 2 years of BIOCHEM course work with summer independent research projects served as the framework for a full scientific publication in Biochemistry (Lukowski et al., 2014). Importantly, the two first authors on this publication were undergraduate students who each took both CHEMBIO and BIOCHEM. Recent cycles of BIOCHEM have also been highly successful, studying distinct, novel mycobacterial serine hydrolases with full structure–function work completed on the narrow substrate specificity of LipW from Mycobacterium marinum (students have prepared a manuscript that is currently submitted for publication) and the broader substrate specificity of the mycobacterial hormone–sensitive lipase LipN from Mycobacterium ulcerans. Individual laboratory experiments from BIOCHEM have also been published with two student coauthors who each took CHEMBIO and BIOCHEM (Johnson et al., 2014a,b). For each BIOCHEM iteration, one BIOCHEM student has also continued the phase 1 classroom research through a summer and school year–independent research project to refine the scientific data collection and extend the scientific investigation.
In addition to direct outputs from the phase 1 structure–function studies of BIOCHEM, phase 2 independent projects have highlighted novel features of serine hydrolases that have continued into independent projects, national scientific meeting presentations for students and faculty, and pending scientific publications (Figure 2, F and G). As an example of this continuation of independent projects, one group applied the kinetic and thermal stability assays from BIOCHEM to understand the effect of divalent metals on the catalytic activity and thermal stability of Rv0045c. Surprisingly, specific divalent metals showed a concentration-dependent effect on the thermal stability and catalytic activity of Rv0045c (Figure 2F; Johnson et al., 2014a; Lukowski et al., 2014). Multiple summer research projects and two senior theses have now refined our understanding of this effect of divalent metals, determining the concentration, structural, and metal dependence of this effect, and these results have been presented at multiple national scientific conferences and are being prepared for publication (see goal 5). The majority of these continued independent research projects were completed by students who had previously taken BIOCHEM. Extensions of independent projects from BIOCHEM are currently ongoing as independent research projects studying the pH dependence, quaternary structure, and three-dimensional structure of mycobacterial hydrolases.
NEURO.
In the NEURO laboratory, students first used fluorescence imaging and RNAi in C. elegans to screen for ubiquitin ligase genes with novel roles in controlling neuromuscular synapse structure (phase 1). Although the impenetrability of the worm cuticle to the fluorogenic probes synthesized in CHEMBIO prevented the incorporation of the esterase–fluorophore pairs into the project design, this initial drawback was taken as a scientific challenge that led to ongoing, collaborative scientific investigation. Two CURE students in collaboration with multiple faculty and two independent research students have continued to investigate this issue and have now collected significant publishable results related to C. elegans serine hydrolases on which all four students are coauthors (the students and faculty [J.R.K. and R.J.J.] are currently writing a paper for submission). All four students involved in this project have presented their CURE-related work at regional and national scientific conferences, three of the four students completed honors theses on this work, and one of the CURE students is also an author on a separate publication based on his additional independent research work on C. elegans neuronal signaling (Kowalski et al., 2014). One additional NEURO student with no prior research experience also went on to do independent research in the instructor’s lab, leading to presentations at local and regional conferences and continuation in a biomedical PhD program (see goal 5).
Following use of the more traditional GFP-tagging approach to visualize neuronal proteins in live worms during the phase 1 RNAi screening, all NEURO groups progressed to screen follow-up and candidate characterization in phase 2 each semester. While the imaging data were intended to document only changes students observed visually in the phase 1 screening, several groups generated imaging and/or behavioral data in phase 2 that were publication or near-publication quality (Figure 2, G–I). For example, several groups tested whether the effects of gene inhibition they observed on synapse structure in the screen correlated with changes in behaviors, such as the ability to swim or “thrash” in liquid (Figure 2H). For genes 1 and 2, the reduced abundance of synaptic vesicles at excitatory cholinergic synapses in RNAi-treated animals correlated with a reduced thrashing ability relative to vector-treated controls (L4440; Figure 2, G and H). Another group found that the reduced synapse density seen with RNAi treatments targeting a different ubiquitin ligase (gene 3) correlated with decreased numbers of cholinergic motor neurons, suggesting effects on neuronal survival or production (Figure 2I). Even for groups without such clean data, more than half generated data on candidates that are sufficiently interesting to warrant additional follow-up studies by students doing independent research projects. To replicate results obtained by groups in one semester without biasing future student groups regarding those genes, genes that showed phenotypes in one semester were included in the lists of candidate genes for future sections. Thus, a number of genes were screened in multiple semesters, and in several cases, the results were replicated or expanded. A handful of promising candidates were also identified that students doing independent research in the instructor’s lab are now characterizing for their roles in neuromuscular signaling.
Overall Course Outcomes.
Together, students in phase 1 of all three CUREs generated novel scientific results, including developing a robust structure–activity library of fluorogenic substrates, dissecting the substrate specificity of multiple mycobacterial serine hydrolases, and identifying novel ubiquitin ligases involved in neural-signaling regulation (Figure 2). As joint faculty, student, and scientific outcomes, these scientific results have been incorporated into joint faculty and student publications and presentations (see goal 5), illustrating the general scientific importance of these CURE investigations. The majority of the CURE results were not immediately ready for publication, but the ability of students in all three CUREs to generate novel publishable or preliminary data supports the findings of these and several other faculty research–driven laboratory courses on a variety of topics (Gardner et al., 2011; Miller et al., 2013; Harvey et al., 2014; Sarmah et al., 2016). The wide range of student-initiated research projects investigated in phase 2 has also started new ongoing research projects in each of the faculty members’ laboratories (Figure 2), reinforcing a previously quantitated benefit to faculty from CURE participation (Sanders et al., 2016; Shortlidge et al., 2016). In addition to incorporation into the research laboratory, CURE results have served as preliminary data for ongoing studies in subsequent iterations of these CUREs, creating a reinforcing cycle and lowering faculty load in future CURE cycles. Similar iterative data-validation approaches were used successfully in previous laboratory courses integrating faculty research projects on the molecular control of python organ regression or neuroanatomical changes in animal models of dyslexia and aging (Gardner et al., 2011; Harvey et al., 2014).
Goal 2: Develop Students’ Experimental Design and Data Analysis Skills
To assess experimental design and data analysis skills of our CURE students, we performed two direct assessments. First, we administered a pair of pre- and postcourse experimental design questions to students in each course (see the Supplemental Material). Each set of questions was specific to the research topic and general experimental approaches used in the course in which it was administered; thus, these open-ended questions were aimed at assessing course content–specific experimental design skills with application to related scenarios. For example, one question each in both BIOCHEM and NEURO asked about the appropriate positive and negative controls needed to accurately interpret and/or troubleshoot particular experimental data. Overall, questions tested students’ abilities to design synthesis schemes and assess product purity in CHEMBIO, to determine the effects of active site mutations on enzyme activity in BIOCHEM, and to select appropriate experimental tests investigating genetic effects on nervous system function versus structure in NEURO. We found that, for five of the six experimental design and data analysis questions (two questions per course), posttest responses were significantly higher than pretest responses (p < 0.005, independent-variables t test; Figure 3A). Only question 2 for BIOCHEM showed no significant increase between the pretest and posttest average course score (p > 0.05, independent-variables t test). The Cronbach’s alpha test for interrater reliability gave scores ≥ 0.9 for both pre- and posttest CHEMBIO questions 1 and 2 and NEURO questions 1 and 2 and for BIOCHEM question 1, indicating excellent reliability (Hallgren, 2012); however, for BIOCHEM question 2, Cronbach’s alpha scores were 0.66 for the pretest and 0.78 for the posttest version, indicating good but still lower reliability. Nevertheless, strong pre- to postcourse improvements seen for questions 1 and 2 in CHEMBIO (59 and 228% increases) and NEURO (198 and 716% increases) and for question 1 in BIOCHEM (40% increase) demonstrate the pre- to postcourse improvement in experimental design skills in students taking each of these courses. As we did not track students who had taken more than one CURE compared with the rest of the population, it is possible that these students performed better on the experimental design questions as a result of the reinforcement of concepts in multiple CUREs; however, we expect that this would have improved both pre- and postcourse scores, potentially canceling any effects. Thus, we think that the measured pre- to postcourse gains were due to student learning and development of the context-specific experimental design skills that were reinforced through their experiments and writing assignments within our CUREs. Overall, the gains in experimental design mirrored the gains in experimental logic reported for a recently modified SEA-PHAGES course-based research experience (Staub et al., 2016) and the content-based gains observed for a variety of other research courses with a range of levels and topics (Shaffer et al., 2010; Campbell et al., 2014; Harvey et al., 2014; Russell et al., 2015). Similar increases in both lower- and higher-order cognitive skills were recently reported for students participating in both CUREs and in independent AREs; interestingly, the pre- to postcourse increases were greater for CURE students, who, on average, began at a lower achievement levels than students in AREs, suggesting the importance of CUREs in narrowing student achievement gaps (Shapiro et al., 2015).
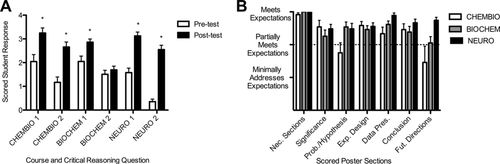
FIGURE 3. Experimental design and data analysis skills improve postcourse. (A) Pre- and postcourse experimental design questions were administered in each course and graded with standard rubrics by all three instructors. Average posttest responses were significantly higher than pretest responses for five questions (*, p < 0.005, independent-variables t test). Cronbach’s alpha scores were ≥ 0.9 for pre- and posttest CHEMBIO and NEURO 1 and 2 and BIOCHEM 1; for BIOCHEM 2, Cronbach’s alpha scores were 0.66 (pre) and 0.78 (post) (n = 22 CHEMOBIO, n = 24 BIOCHEM, n = 33 NEURO). (B) Student groups prepared posters on their research. Each poster was rated in seven areas by all three instructors using a standard rubric. Average scores for each area were determined for each course. Cronbach’s alpha tests showed fair to excellent interrater reliability (necessary sections = 1.0 [excellent]; significance = 0.805 [excellent]; problem/hypothesis = 0.796 [excellent]; experimental design = 0.588 [fair]; data presentation = 0.713 [good]; conclusions = 0.805 [excellent]; future directions = 0.929 [excellent]; n = 8 CHEMBIO, n = 12 BIOCHEM, n = 10 NEURO; Hallgren, 2012). Error bars show ± SEM.
As a second, independent measure of students’ mastery of scientific skills related to all aspects of experimental design and scientific communication, we rated the final posters that groups of students prepared on their projects in each course for competency in each of seven areas: 1) necessary sections present, 2) significance of project, 3) problem/hypothesis statement, 4) experimental design, 5) data presentation, 6) conclusions, and 7) future directions (Figure 3B, Supplemental Material 3). While all posters did not fully meet expectations in any one category, we found that groups in all three courses at least partially met expectations in all categories, except for the problem/hypothesis and future directions areas in CHEMBIO (Figure 3B), demonstrating general competence in these varied aspects of experimental design and data analysis and suggesting overall competency of the students in understanding and presenting scientific research information in this format. Such participation of students in “authentic scientific communication” activities is one of the core recommendations for successful development of a research-based course (Kloser et al., 2011). Students in all three courses received feedback on drafts of their posters and presentations and on scientific writing exercises (e.g., manuscripts and/or project proposals) before submitting their final versions, again meeting suggested criteria for maximizing student learning gains from research-based courses (Kloser et al., 2011).
Goal 3: Promote Positive Student Attitudes about Science and Perceptions of Learning Gains
Third, we assessed student attitudes about science and students’ perceptions of their own learning gains related to their research skills using the CURE survey, which uses pre- and postcourse questions to measure students’ perceived learning gains in courses with significant research components (Lopatto, 2004, 2007; Lopatto et al., 2014). These data are compared with CURE survey results from students in other classroom-based research courses and in SUREs. We selected groups of CURE questions related to research preparation and confidence (six questions), scientific communication skills (three questions), data analysis and lab skills (three questions), and interest in science and research (two questions). For all 14 questions in all four categories, students in CHEMBIO, BIOCHEM, and NEURO scored at or above the combined mean of all CURE and SURE students. Particular areas of strength in which our students’ scores were notably above the CURE + SURE average include “self-confidence,” “readiness for more demanding research,” “understanding the research process,” “skill in scientific writing,” “good way of learning about the scientific process,” and “positive effect on interest in science.” These skills span all four areas assessed, indicating the perceived gains our students made in all of these areas—gains equivalent to or greater than those made by students in similar courses or participating in various summer and course-based research programs (Figure 4; Lopatto, 2004, 2007; Lopatto et al., 2014). Gains were particularly notable in areas related to self-confidence, preparation for more demanding research, and interest in scientific research, corroborating the improvements in experimental design skills and the competence in experimental design and analysis on the posters that we measured directly. These gains also correlate with increases in student responses to the CURE survey or similar attitudinal surveys regarding students’ perceptions of their readiness for research, confidence in skills related to performing scientific research, understanding of how scientists think, and understanding of the research process reported for other research-based courses (Miller et al., 2013; Harvey et al., 2014; Russell et al., 2015).
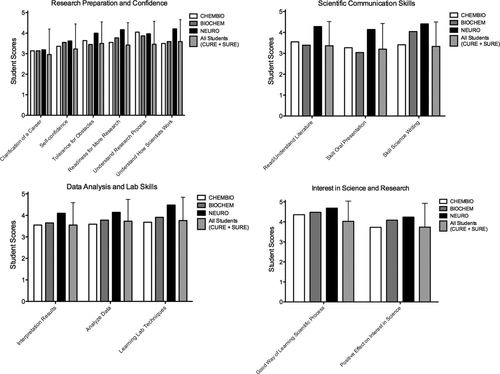
FIGURE 4. Students perceive gains in research skills and exhibit confidence and interest in research. Student attitudes about science and perceptions of learning gains related to their research skills were assessed using the CURE survey (Lopatto, 2004, 2007; Lopatto et al., 2014). Results were compared with CURE survey results from students in other CUREs and in SUREs. We selected groups of CURE questions related to research preparation and confidence, scientific communication skills, data analysis and lab skills, and interest in science and research. For all questions shown, students in each course scored at or above the combined mean of all CURE and SURE students. Error bars show ± SD of the mean of “all students (CURE + SURE).” (CHEMBIO 2012–2014: n = 22 students pre, n = 22 post; BIOCHEM 2013–2015: n = 22 pre, n = 23 post, NEURO 2013–2015: n = 35 pre, n = 29 post).
Together, data from our direct assessments of students’ experimental design skills and our indirect assessments of student attitudes and perceptions of their own learning gains support the success of each of our CUREs in promoting scientific skills and positive attitudes about science—critical, yet often poorly documented outcomes of CUREs (Corwin et al., 2015; Linn et al., 2015). Student comments on open-ended, end-of-semester course evaluations support our CURE survey results (Supplemental Table S2), as students noted how much they enjoyed the in-depth focus on a single project, which allowed them to become “very well-versed” and “more accomplished,” to learn “how to solve problems,” to have “a greater understanding of specific results,” “allowed for more creativity,” and helped them “fe[el] like we were doing important work.” Students also mentioned how much they enjoyed having their own projects and designing their own experiments and were “really invested in coming back,” suggesting strong feelings of project ownership, a validated outcome from CURE participation (Corwin et al., 2015). In the future, we hope to quantify this ownership using recently validated and published methods (Hanauer and Dolan, 2014).
Goal 4: Promote Student Retention in STEM Disciplines
On the basis of previous CURE studies (Auchincloss et al., 2014; Brownell et al., 2015; Corwin et al., 2015), we expected that participation in CUREs would promote student interest in scientific research and STEM disciplines. We found that an average of 74.7% of the 79 total students enrolled in at least one of our three research-based courses also participated in independent UREs (Table 2). This is significantly above the 40% average research participation for the biological sciences and chemistry departments at Butler and is similar to the 80–90% of students who sought research positions following their experiences in the research-intensive Python Project lab course at the University of Colorado (Harvey et al., 2014). Additionally, nearly one-third (32.9%) of the 79 enrolled students completed an honors thesis, which is also significantly above the 9% average of STEM students who completed a thesis at Butler University from 2003 to 2012 (Howes and Wilson, 2015).
| Course | Undergraduate research (%) | Undergraduate thesis (%) | STEM graduate school (%)a | STEM professional school (%)b | STEM employment (%) | Total students |
|---|---|---|---|---|---|---|
| CHEMBIO | 81.8 | 40.9 | 50.0 | 50.0 | 0.0 | 22 |
| BIOCHEM | 75.0 | 33.3 | 52.6 | 42.1 | 5.3 | 24 |
| NEURO | 69.7 | 27.3 | 33.3 | 60.0 | 0.0 | 33 |
| Total | 74.7 | 32.9 | 43.3 | 52.2 | 1.5 | 79 |
The increased level of participation in UREs by our CURE students is an exciting outcome of our CURE courses. After 3 years of CURE implementation, this increased level of URE participation is, however, reaching the maximum capacity of our departments to provide high-quality independent research opportunities. Yet, based on our assessments of student gains in experimental design (Figure 3) and student perceptions of learning gains (Figure 4), our CUREs provide students with the measureable outcomes of UREs without the increased strain on limited departmental resources. Continued implementation of these CUREs will thus allow us to provide authentic research experiences to the greatest number of students. Thus, CUREs can generate student populations better prepared to engage in future independent research and allow more students access to real-world, student-driven research experiences not available in the traditional laboratory classroom.
Regarding their postgraduate career goals, slightly less than half (43.4%) of students are pursuing or intend to pursue STEM-related graduate degrees (e.g., master’s or PhD programs in chemistry, biology, or psychology), while slightly more than half (52.2%) are pursuing or intend to pursue training in STEM-related professional schools (e.g., medicine, dentistry, veterinary medicine, physical therapy, physician assisting, clinical psychology, and genetic counseling). One student (1.5%) is pursuing STEM-related employment without further education. In total, 97% of all the students in each of our three research-intensive courses between 2012 and 2015 intend to continue in STEM-related careers. Given the small size of both the Department of Chemistry and the Department of Biological Sciences at Butler, we do not have data on sufficient numbers of students to make definitive comparisons between the retention rate of CURE students and students outside this cohort. However, a similarly high level of retention in STEM disciplines was seen for students participating in introductory-level CUREs on zebrafish development, in which more than 80% of students intended to continue in the biomedical field at the end of the course (Sarmah et al., 2016) or in one of several integrated CUREs at Georgia Gwinnett College (Russell et al., 2015).
While we do not have sufficient comparative data to determine whether our students would have chosen these STEM career trajectories even in the absence of enrolling in our courses, we have additional qualitative evidence to suggest that participation in these courses helped students clarify their career goals with regard to research. Although we know that multiple students participated in undergraduate research before enrollment in our CUREs, we know of students from NEURO and from CHEMBIO who started independent undergraduate research after completing the CURE and are now enrolled in a biomedical or chemistry PhD program specifically because of their research experiences in these courses. Although there is likely some self-selection of research-oriented students into our courses, these data are consistent with previous reports of the positive impact of research on students’ likelihood of enrolling in STEM graduate programs and overall persistence in science (Eagan et al., 2013; Corwin et al., 2015). A recent study assessing both pre- and postcourse career intent found little change in the overall number of students interested in pursuing STEM careers but did see a ∼15% increase in the number of students intending to pursue a master’s or PhD at the end of the research-based course with concomitant decreases in the number planning to pursue professional degrees in medicine or pharmacy (Harvey et al., 2014). Conversely, we know of students from NEURO and BIOCHEM for whom their CURE experience helped them to decide that the research field for the course is not the ideal career path for them and solidified their desire to pursue professional studies in a different field. Thus, student exposure to our CUREs later in their academic careers was still able to impact their feelings toward research and career decisions, underscoring the importance of integrating this type of course into undergraduate curricula at all levels.
Goal 5: Promote Faculty Research Productivity
In addition to student benefits, we predicted that this series of CUREs would increase the research productivity of participating faculty members, as each faculty member was able to incorporate his/her own research into the course and train a population of students who would be motivated and capable of engaging in independent laboratory research. To test this prediction, we measured faculty research productivity before and after CURE implementation by determining the mean number of research students mentored in faculty members’ labs per semester and the total number of peer-reviewed research publications and the number of presentations for each faculty member. We found increases in the combined data from all three faculty members and for each individual faculty member for all three productivity parameters during the 3-year period following initial implementation of the courses (Summer 2013–Spring 2016) compared with the 3 years before course implementation (Fall 2009–Summer 2012; Table 3). The mean number of research students mentored per semester increased 1.35-fold from the pre- to postcourse period, with individual faculty member increases ranging from 1.14- to 1.69-fold. The increase in the combined number of research students per semester for all faculty members was statistically significant (p = 0.014, Kruskal-Wallis nonparametric test), as was the increase seen for faculty member 1 (J.R.K.) alone (p < 0.05, two-tailed unpaired t test, assuming unequal variance). In addition, there was a 3-fold increase in the number of combined publications and a 2.4-fold increase in presentations from the precourse to the postcourse period, with a range of 0.5-fold to 7-fold increases in individual faculty member publications and a range of 2-fold to 3.3-fold increases in individual faculty member presentations. It is important to note that some increase in absolute numbers of research students and publications/presentations may also be due to the fact that two of the three faculty (J.R.K. and R.J.J.) were recently hired during the 3 years before implementation of the courses and were tenured by the end of the 3-year postcourse period. Irrespective of faculty position, however, these faculty benefits likely would not have resulted from continued participation in courses in which students performed only traditional laboratory experiments. Moreover, we think that the large increases in productivity seen for the newly hired faculty (1 and 2, J.R.K. and R.J.J.), the number of publications directly related to the CURE research, and the fact that the senior faculty member (3 , G.C.H.) still saw a substantial increase in productivity provide compelling support that some of the increased faculty productivity was a direct result of involvement in these CUREs. Overall, our total publication counts, in both science and science education, were above the 1.32 average publication outcomes measured for 38 CURE active faculty (Shortlidge et al., 2016). Furthermore, publication numbers are underrepresented in this table, as several faculty members have additional CURE-related publications in various stages of preparation and the data presented do not account for lab-connected outcomes that have been incorporated into successful external grant applications.
| Research students per semester | Publications | Publications with students (CURE) | Presentations | Presentations with students (CURE) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Faculty member | PRE | POST | PRE | POST | PRE | POST | PRE | POST | PRE | POST |
| 1 | 4.1 ± 1.9 | 6.6 ± 1.4* | 1 | 2 | 0 | 2 (1 + 1 in prep) | 13 | 43 | 13 | 41 (7) |
| 2 | 6.4 ± 3.7 | 7.3 ± 2.4 | 1 | 7 | 1 | 7 (6) | 29 | 58 | 27 | 57 (35) |
| 3 | 2.2 ± 1.2 | 3.6 ± 1.8 | 2 | 3 | 0 | 3 (3 + 1 in prep) | 6 | 18 | 5 | 17 (16) |
| Total | 12.8 ± 4.8 | 17.4 ± 4.8** | 4 | 12 | 1 | 12 (10 + 2 in prep) | 48 | 117 | 48 | 115 (58) |
The direct overlap between the faculty members’ research and the CUREs was likely a supporting factor for our high number of publications. Nearly all of the faculty publications and presentations involved student coauthors both before and after implementation of the CUREs (Table 3). This is likely reflective of the commitment of these faculty members, and Butler University as a whole, to promoting faculty–student research partnerships. Thus, implementation of our CUREs correlated not with an increase in the percentage in faculty publications involving students but with an enhancement of the overall research productivity of each faculty member. Moreover, between 50 and 100% (75% overall) of all faculty–student coauthored publications and between 17 and 94% (50% overall) of faculty–student coauthored presentations in the post-CURE period directly resulted from research related to work done in the CUREs (Table 3), suggesting that integration of faculty research into these courses had a direct and positive impact on faculty research productivity. This overlap of teaching and research matches with the synergism between teaching and research observed at other liberal arts institutions, reinforcing a major benefit identified by other CURE faculty from independent CUREs (Brownell and Kloser, 2015; Shortlidge et al., 2016). Such benefits require that faculty research be easily integrated in a CURE format, which was one impediment mentioned by other CURE faculty (Lopatto et al., 2014; Shortlidge et al., 2016). The success of CHEMBIO and BIOCHEM, in particular, in producing publication and presentation outcomes may be related to the direct overlap between scientific goals from the two courses in which intellectual and experimental continuity between these two courses amplified faculty benefits. Finally, in terms of student participation, each faculty member directly assimilated multiple students from one of the CUREs into his/her research lab and can attest to the increased intellectual and experimental preparation of the CURE students for this work.
Overall, from our experience as CURE faculty, this teaching and learning model was beneficial for both student learning and engagement, as well as for the productivity of faculty with significant commitments to both teaching and research (Shortlidge et al., 2016). Major indirect benefits of the CUREs for faculty have been the development of new research directions and self-continuation of the research courses through preliminary data from students’ independent projects, illustrating broadened faculty research interests and directions from CURE participation (Shortlidge et al., 2016). These findings lend credence to the suggestions made in several recent reports that incorporation of faculty research into classroom laboratories may help overcome the limits of time and institutional support for faculty balancing significant research and teaching duties (Darden, 2003; Kloser et al., 2011; Miller et al., 2013; Lopatto et al., 2014). Much like the benefits ascribed to a strong central support system for implementation of multi-institution CUREs (Lopatto et al., 2014), our experiences with three collaboratively designed CUREs promoted shared research and pedagogical resources that eased the burden on individual faculty members running each course. These benefits also match with the reimagining of teaching and research proposed by Boyer (1990), so that teaching is not merely a teaching load but rather an integrative mixture of teaching and research that fuels ongoing advancement in teaching and research. Further, the benefits of our collaborative CUREs extend beyond the context of these courses, as we have experienced overall increased interdepartmental collaboration, perhaps the best example of which is the plan currently underway for shared interdepartmental molecular life science research and teaching space in our institution’s new science facility. We anticipate additional collaborations in teaching and research will result from our future proximity and shared interests. Additionally, as a result of the success of our efforts to incorporate independent research projects in our collaborative CUREs and the growing interest of other faculty in developing research-based lab courses, the Department of Biological Sciences recently adopted a policy in which upper-division lab courses will incorporate some independent project work.
Challenges to Implementation
Mirroring previous CUREs, our faculty and student benefits from CURE participation were not without faculty challenges to implementation and successful outcomes (Kloser et al., 2011; Lopatto et al., 2014; Shortlidge et al., 2016). For our CURE design, faculty research directions had to be realigned to fit more closely to a standard laboratory session, to match with the budgetary framework available for the courses, and to maximize student success on course projects (Shortlidge et al., 2016). One approach we used to overcome these hurdles and to minimize initial faculty input was to design each individual CURE laboratory with a parallel structure and framework to previous CURE laboratories, for example, protein structure manipulation by mutagenesis and parallel synthesis for library construction (Bailey, 2009; Knutson et al., 2010a; Scott et al., 2015). These overall structures were tweaked to incorporate our specific experimental methods and realigned around the central themes of mycobacterial hydrolases and neural signaling. This realignment required more time, logistical support, and financial commitment than traditional laboratories; however, the use of common molecular and cellular equipment (PCR machines, gel-electrophoresis systems, bacterial cell culture materials, and microscopes) in all three CUREs has facilitated the development of other research-based courses on different topics within the molecular sciences.
One additional concern of particular note is that, while students felt our CUREs were of great value in terms of their scientific training, multiple students voiced frustration with the amount of effort required for these courses relative to the amount of credits they received (Supplemental Table S2). This concern is valid, as students were asked to do significant writing, reading of scientific literature, poster preparation, and some lab work outside the scheduled lab times, yet traditionally students and faculty at many institutions like ours receive only half-credit for time spent in the lab. Thus, while we have worked to minimize the lab time required outside scheduled times or allowed group members to split responsibility for completing assigned lab tasks, the issue of workload relative to course credits is an ongoing challenge to implementation. Ultimately, our student and faculty outcomes and our own personal enjoyment strongly argue that CURE design and implementation are worth the time, effort, and resources invested, and through our collaborative model, we worked to minimize strain on faculty and student workloads.
Finally, small-scale CUREs such as ours also raise the question of scalability when considering the transferability of these courses to other institutions with higher numbers of students per course. While we acknowledge that the cost of reagents and access to equipment may limit the possibility of our specific CUREs being implemented in courses with multiple sections with larger enrollments, the design of each of our CUREs is such that the research project can be modified to accommodate a range of institutions. For example, in CHEMBIO, students could work in groups to synthesize novel fluorogenic derivatives or groups could work in parallel to construct identical structural modifications on different fluorophore scaffolds. In BIOCHEM, the number of serine hydrolase enzymes and the range of selected mutations of those enzymes could be greatly expanded, greater redundancy could be covered in student variants, or a single-site saturation mutagenesis reaction could be performed to create a library for the class to collaboratively analyze. In NEURO, a larger number (>600) of ubiquitin ligase genes are available for student groups to screen or rescreen for their effects on synapse structure and function, students could screen for the effects of the same ubiquitin ligase genes on excitatory versus inhibitory synapses, or the effects of ubiquitin ligases on behavior rather than synapse structure could be used as a simpler initial screen. Overall, although there would be a cost to scaling up any of these CUREs, the research projects and the reagents used are flexible and transferable for use in different institutions.
CONCLUSIONS
In conclusion, we designed and implemented a collaborative series of upper-division CUREs in chemical biology, biochemistry, and neurobiology that involve students in novel research projects addressing questions of broad scientific importance that are directly tied to faculty research interests and expertise. Our combined assessments indicate that both students and faculty benefit from participation in these courses, as students demonstrated increased experimental design and data analysis skills and positive attitudes about research and their own scientific skills and retention in STEM-related disciplines. Faculty also benefited, as we noted increased research productivity, increased student participation in our research labs, and increased collaboration. This course design and implementation present a cohesive CURE model of how CUREs involving collaboration among multiple faculty across disparate scientific fields and departments can be linked across a university curriculum to provide significant learning and career benefits to an interdisciplinary population of students and faculty at a single institution.
With the success of the course design, implementation, and significant student–faculty benefits, we envision future expansion and improvements in course design and assessment. In addition to strengthening current links, we envision expanding this series to include other collaborative upper-level courses such as bioinformatics, molecular biology, or cell biology. We would also like to add CUREs at lower levels of the curriculum that students take before taking these stand-alone electives, and more open-ended, research-based lab activities are already being implemented throughout our curricula since the initiation of our CUREs. Ideally, these courses would also be collaborative, such that students could continue to experience the multifaceted approach required to solve global scientific problems and to further strengthen research and teaching ties between the science departments.
While we would gladly support the implementation of these specific CUREs at other institutions, including modifications to better suit specific courses and institutions, we think that our CUREs more generally provide a model that could be used by faculty collaboratively teaching other sets of courses on any number of different topics. Our experience and analysis of these CUREs strongly support the emerging body of evidence arguing for the substantial long-term benefits of reimagining laboratory education to bridge the disconnect between teaching and research.
ACKNOWLEDGMENTS
We thank Dr. Richard Nass (Indiana University School of Medicine) and Dr. Luke Lavis (Howard Hughes Medical Institute–Janelia Farms) for experimental reagents, Dr. Anne Wilson (Butler University) for her assistance with rubric verification and data analysis, Dr. Lars Dreier (UCLA) for creating the original C. elegans neuron image in Figure 1, and Dr. David Lopatto and Leslie Jaworski (Grinnell College) for their assistance with data collection and analysis for the CURE survey, which is supported by the Howard Hughes Medical Institute. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We also thank all of the Butler University undergraduate students in CHEMBIO, BIOCHEM, and NEURO from Fall 2012 to Spring 2015 for their hard work and feedback for these courses. This project was funded by National Science Foundation-TUES award (DUE-1140626) to R.J.J., J.R.K., and G.C.H.



