Using Next-Generation Sequencing to Explore Genetics and Race in the High School Classroom
Abstract
With the development of new sequencing and bioinformatics technologies, concepts relating to personal genomics play an increasingly important role in our society. To promote interest and understanding of sequencing and bioinformatics in the high school classroom, we developed and implemented a laboratory-based teaching module called “The Genetics of Race.” This module uses the topic of race to engage students with sequencing and genetics. In the experimental portion of this module, students isolate their own mitochondrial DNA using standard biotechnology techniques and collect next-generation sequencing data to determine which of their classmates are most and least genetically similar to themselves. We evaluated the efficacy of this module by administering a pretest/posttest evaluation to measure student knowledge related to sequencing and bioinformatics, and we also conducted a survey at the conclusion of the module to assess student attitudes. Upon completion of our Genetics of Race module, students demonstrated significant learning gains, with lower-performing students obtaining the highest gains, and developed more positive attitudes toward scientific research.
INTRODUCTION
In recent years, next-generation sequencing (NGS) technology has revolutionized the ability to extract massive amounts of data from biological samples (Shendure and Aiden, 2012). Owing to the combination of NGS technology and new bioinformatics tools, genetic sequencing has expanded into many applications that influence our everyday lives, such as healthcare and personalized genomics. The expanding availability and utility of genetic information make it increasingly important for the general public to have a level of comfort with genetic information and an understanding of how to interpret this information. For many individuals, the genetics curriculum mandated in high schools is the most in-depth curriculum they receive. However, current high school curricula often have limited coverage of genetics and bioinformatics despite the growing importance of these topics (Wefer and Sheppard, 2008; Dougherty et al., 2011). As a result, students miss out on recent technological developments that would convey the significant and personally relevant role that genetics increasingly plays in our health and lives.
To address this disconnect between high school curricula and current uses of sequencing and bioinformatics, we have developed laboratory-based modules to bring these modern genetics techniques into high school classrooms. Through a National Institutes of Health (NIH) Science Education Partnership Award, we established an educational outreach program, Bioinformatics Inquiry through Sequencing (BioSeq, 2016), including a fully functioning educational sequencing center equipped with the latest NGS technology. Making use of this educational sequencing center, we designed modules around personally relevant topics that would involve students in processing their own samples and analyzing their own data. We adopted an inquiry format for these modules, encouraging students to interpret data and justify their own conclusions to answer an open-ended research question. To further promote student engagement, we involved undergraduate students as “near-peer” mentors both in the development of the laboratory-based curricula and in leading the modules during classroom implementation.
In this paper, we describe the implementation and initial evaluation of our “The Genetics of Race” laboratory module. In this module, students make predictions about genetic similarity (“To whom am I most genetically similar in this classroom?”), which are often associated with perceived racial identifiers. We approached the potentially charged topic of race as a hook for this experiment in order to connect the issues of modern society with current methods in genetics. While the concept of race has a troubled history with biology (Yudell et al., 2016), the development of modern sequencing and bioinformatics techniques has given us more sophisticated tools for exploring race and genetics (Bamshad et al., 2004; Tishkoff and Kidd, 2004; Berg et al., 2005; Bonham et al., 2005). Our module provides access to these techniques, enabling students to explore their own genetic similarity with their classmates. While our informal observations suggest that the Genetics of Race module prompts students to examine their assumptions about the link between genetics and race, we do not aim to change student attitudes about race as a learning goal in our implementation. Rather, we use race as a hook to engage students and emphasize the personal relevance of sequencing.
We sequence student samples using NGS technology to deliver a unique educational experience with the latest sequencing advances. However, NGS requires specialized equipment and can be expensive (up to $2800 per implementation of the module). If NGS technology is difficult to access or cost-prohibitive, traditional Sanger sequencing can be used as an alternative method. Costs for both NGS and Sanger sequencing are reviewed in more detail in the Discussion and in the Supplemental Material.
METHODS
Module Development
Educators interested in using our instructional materials may visit the BioSeq website at http://ase.tufts.edu/chemistry/walt/sepa/geneticsofrace.html or contact the corresponding author.
Our Genetics of Race laboratory module was inspired by classroom experiments carried out at Clemson University (Kosinski et al., 2008) and the DNA Learning Center at Cold Spring Harbor Laboratory, as featured in the PBS program Race: The Power of an Illusion (California Newsreel, 2003; Public Broadcasting Service, 2003). In our module, we highlighted NGS technology to harness student excitement about recent innovations in genetics technology, and we developed laboratory procedures specifically to be compatible with our NGS laboratory. Curricular materials were designed for the target audience of standard-track biology and biotechnology high school classes. Tufts undergraduate students played an active role in designing both laboratory and classroom materials for the module.
Undergraduate students also were responsible for leading classroom visits as we implemented our module at local schools. We first implemented the Genetics of Race module in several high school biotechnology classes. A summary of the Genetics of Race module is shown in Table 1. In addition to the five main student-led lessons, we also visited the classrooms to distribute consent forms and administer assessments. Furthermore, we hosted a field trip to the BioSeq educational sequencing center so students could visit a genuine research laboratory and see where their samples were sequenced. The entire module was spread over 4 weeks to allow for adequate sample processing and sequencing time. Other than the lab tour and final sample processing and sequencing steps, both of which were performed at our sequencing center, all materials and sample processing steps were presented or performed in the classroom.
| Classroom visit | Description |
|---|---|
| Lesson 1: Introduction | Clarify definitions related to race and ancestry Introduce the genetics of race experiment |
| Lesson 2: Sample collection | Student extraction of DNA from their buccal swab samples Transport samples to Tufts University for final processing and sequencing |
| Lesson 3: Sequencing by synthesis | Review DNA synthesis Discuss the modifications to DNA synthesis that enable modern sequencing technologies |
| Lesson 4: Data analysis | Analyze DNA sequence information, bioinformatics intro Practice analyzing example DNA sequences |
| Lesson 5: Results and discussion | Distribute student results Share and compare conclusions |
Module Implementation
Lesson 1: Introduction to the Genetics of Race Experiment.
Because race serves as a hook for our module, we begin the implementation by providing the background and research questions that relate to race. The first class begins by asking the students to define the terms “race” and “ancestry.” Student answers often conflate the two; our initial discussion prepares students to explain the difference between “race” (socially defined, corresponds to a small set of perceived observable traits) and “ancestry” (can be scientifically defined, corresponds to genetics). We then ask students whether DNA could be used as an identifier of someone’s race. To explore this possibility, we introduce mitochondrial DNA and preview the sample collection procedure that will be carried out in the next lesson. We note that mitochondrial DNA is inherited only from the maternal side and does not undergo recombination. In addition, mitochondrial DNA is a good target for a high school experiment, because there is a relatively high copy number present in each cell, making it easier to obtain an amount sufficient for sequencing (Salas et al., 2000). At the conclusion of the first lesson, students also record their initial predictions about the classmates to whom they expect to be most and least genetically similar based on a comparison of mitochondrial DNA sequences. Students were prompted to justify their predictions. We collected these predictions, which served as the students’ initial hypotheses, to return to the students at the conclusion of the experiment.
Lesson 2: Collecting DNA Samples.
The laboratory component of the module requires students to collect and extract their own mitochondrial DNA. To begin sample collection, students swab the inside of their cheek to collect buccal cells. Students then submerge the cells on the swab in an extraction buffer and lyse the cells with heat. This step exposes the DNA in preparation for subsequent amplification by polymerase chain reaction (PCR). The sample-collection procedure requires micropipettes and tips, sterile foam-tipped swabs, vortexers, a hot-water bath, and reagents from a commercially available kit for DNA extraction. We provide these materials as needed; a materials list and procedure are provided in the Supplemental Material. Once students complete the DNA isolation steps, their samples are transported to our educational sequencing lab at Tufts University for processing, with a subsequent workflow as described in Figure 1. These laboratory and data analysis steps are described in the Supplemental Material.
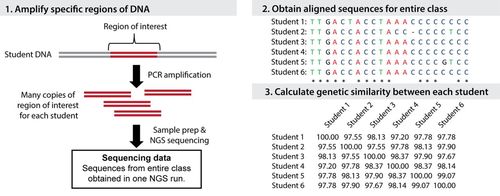
FIGURE 1. Summary of sample processing and data analysis steps for the Genetics of Race experiment. These steps are performed after students obtain buccal swabs and extract their DNA in the classroom. Section 1 illustrates the amplification of the desired region of mitochondrial DNA for each student sample followed by further sample preparation and sequencing. Section 2 shows the alignment of multiple student sequences. In this example, only a short fragment of each student sequence is shown. Section 3 shows the calculated genetic similarity between each student based on the alignment in section 2. The most and least similar values are provided to each student. These steps are also discussed in the Supplemental Material.
Classroom Visit to Educational Sequencing Facility.
To permit the high school students to see the entire experimental workflow, we host a group of students on a field trip to our sequencing laboratory. Students learn about the instrumentation that is used for preparing samples for sequencing, including tools for DNA manipulation and characterization, as well as the sequencer itself. They also perform common laboratory techniques such as gel electrophoresis and DNA quantification. Students participate in making a short video that is presented to all the classes, so students who were unable to join the field trip can also see the laboratory and experimental process.
Lesson 3: Sequencing by Synthesis.
To understand the concept of DNA sequencing by synthesis, students first review how DNA synthesis occurs in biological systems. Using natural DNA synthesis as a starting point, students then investigate how this process can be modified to enable sequencing by synthesis. To demonstrate the technical process of sequencing, students participate in a role-playing activity in which they take on roles portraying the components of a modern sequencing protocol (such as DNA polymerase and free-floating nucleotides) and act out the process of sequencing a short unknown DNA sequence.
Lesson 4: Data Analysis.
With this lesson, students work with simulated data to master the general bioinformatics concepts and the data analysis workflow performed on their samples. First, to demonstrate the concept of DNA assembly, students work with fragmented song lyrics with the goal of reassembling them into the original song, either “resequencing” (using a lyric sheet as a reference) or “de novo” (without any reference). Next, students practice DNA alignment and quantifying percent similarity between different sequences. Once students are comfortable with this concept, each student is then provided with a short sequence of example DNA to compare with those of their classmates, carrying out the process of multiple sequence alignment. They use this activity to construct a similarity matrix for the mock data and determine the most and least genetically similar pairs of samples in the example data set.
Lesson 5: Results and Discussion.
Students receive result reports with four pieces of information: the identities of their classmates with the most and least genetic similarity and the percent genetic similarity associated with each of these comparisons (Figure 2). Before we distribute the results, we remind students to respect the privacy of their classmates. In the event that data are not available for a student, we provide him or her with an alternative result from BioSeq personnel or their teacher. As with the standard results, these alternative results also include a comparison between the sampled individual and the other students in the classroom.

FIGURE 2. Sample results as received by students in the Genetics of Race experiment. Students who do not work with their own samples or students whose samples could not be sequenced due to technical difficulties, instead receive results corresponding to BioSeq personnel or their teacher. These results will indicate the classmates with the most and least genetic similarity based solely on the two short regions of mitochondrial DNA sequenced in the experiment. If more than one student is tied for highest or lowest genetic similarity, the names of all tied students will be listed.
Upon receiving their results, students review the research question for the experiment, the experimental workflow, and their initial predictions. We ask students to consider the limitations of the data, including the major limitation that mitochondrial DNA only reflects maternal lineage and does not capture the entire ancestry of an individual. After students discuss their individual results, the entire class collaborates to determine how many students were correct about their initial hypotheses. Students compare how the class-wide results matched their expectations based on perceived racial similarities. We conclude the lesson by exploring the question “How could this experiment have been completed without sequencing?” and brainstorming ideas for other experiments that can be completed with sequencing and bioinformatics.
Privacy and Sensitivity Concerns
We worked closely with our institutional review board (IRB) to navigate concerns with the use of personal genetic information by minors. Student privacy was protected throughout this study by anonymizing student samples and data using randomized ID numbers from the start of the experiment. The information linking ID numbers to student identities was only known to the BioSeq study coordinator, who replaced the ID numbers with actual student names immediately before returning the results to the students.
The students’ own DNA sequences were not revealed to them as an additional precautionary measure to protect against health and family privacy concerns. This restriction ensured that students could not draw unwarranted conclusions about their family history or health based on their DNA sequences. We also sought to minimize potential health implications by sequencing only small noncoding portions of the mitochondrial DNA, regions that do not code for any physical traits but are often used to track migration and genetic variation between human populations.
Despite these precautions, the use of actual genetic similarity data could potentially lead to sensitive situations. We anticipated two likely scenarios and planned for how to address them should they arise. The first scenario involved two maternally related siblings who do not have 100% mitochondrial DNA similarity. (Because mitochondrial DNA is only passed on through the mother, paternal blood relations do not play a role in mitochondrial DNA similarity.) We can address this concern by explaining how, due to the high rate of mutations in the hypervariable region, even the same person could have multiple different sequences for their own mitochondrial DNA (a common condition known as heteroplasmy; Ramos et al., 2013). Point mutations in the hypervariable regions have also been observed to increase with age (Kennedy et al., 2013). Thus, it is possible for a mother to develop a mutation and pass it on to just one child, or for the child to develop a mutation that causes a reduction in similarity to a sibling. The second scenario would arise if two unrelated students obtain a 100% mitochondrial DNA match. We can explain this outcome by emphasizing that we are only sequencing a relatively short DNA region, and we capture only a tiny fraction of the genetic variation between individuals. A result of 100% genetic similarity does not indicate a direct family relationship, although it suggests a shared common ancestor at some point.
We also consider the possibility that some students’ results may confirm racial stereotypes. We use this potential outcome as an opportunity to discuss the limitations of our study: we sequenced an extremely small portion of the genome, and our sample size is small. We remind students that we should consider all results from the class and share results from larger studies that indicate the loose and complicated relationship between genetics and race.
Assessment Tools
Assessment Design and Methods.
Our assessments sought to measure the influence of our intervention on student knowledge and attitudes regarding sequencing and bioinformatics. We evaluated the effects on students who participated in our program using a knowledge test and an attitudinal survey (both provided in the Supplemental Material) prepared in collaboration with Davis Square Research Associates (DSRA). The knowledge test was developed by BioSeq personnel and consists of 10 multiple-choice questions, querying knowledge across the domains of 1) sequencing and ethics and 2) bioinformatics (five questions for each domain). The questions can also be loosely categorized into three functional categories: “details of sequencing” (four questions), “common techniques and tools” (four questions), and “interpretations and limitations of sequencing results” (two questions). This instrument was administered before and after the module content was implemented. Knowledge tests were scored by BioSeq personnel and the data were analyzed by DSRA.
The attitudinal survey was jointly developed by DSRA and BioSeq personnel. The survey comprises three domains: knowledge, skills, and self-efficacy around the study of genetics. This survey is administered online after the intervention and asks students to self-report on their attitudes both pre- and postintervention. Data were gathered directly by DSRA. Students used their anonymized ID numbers for both the knowledge test and attitudinal survey. In the analysis of assessment results, effect sizes (as shown in Tables 2 and 3) were determined by Cohen’s d, and reliability (as shown in Table 3) was calculated using Cronbach’s alpha statistic. We coordinated with our IRB throughout the design of all assessment tools.
| Test | Mean pretest score | Mean posttest score | % Gain | Effect size (Cohen’s d) |
|---|---|---|---|---|
| Sequencing and ethics (maximum = 5) | 1.81 | 3.31 | 82.87 | 1.21 |
| Bioinformatics (maximum = 5) | 1.06 | 2.78 | 162.26 | 1.73 |
| Test totals (maximum = 10) | 2.88 | 6.09 | 111.46 | 1.72 |
| Construct | Mean gain | SD | % Gain | Effect size (Cohen’s d) | Reliability (Cronbach’s α) |
|---|---|---|---|---|---|
| Knowledge self-assessment | 11.56 | 5.68 | 89 | 2.88 | 0.884 |
| Skills self-assessment | 7.83 | 5.20 | 69 | 2.13 | 0.771 |
| Self-efficacy | 10.30 | 7.89 | 47 | 1.85 | 0.954 |
| Total | 29.70 | 16.24 | 64 | 2.59 | 0.955 |
Population Information.
The population for the data reported here included 68 high school students enrolled in four biotechnology classes at the same school. The student makeup was diverse both racially (63% nonwhite) and ethnically (40% Hispanic). More detailed demographic information is provided in the Supplemental Material.
RESULTS
Knowledge Test
Based on the pretest and posttest scores, high school students who participated in the Genetics of Race module showed strong learning gains, as shown in Table 2 and Figure 3. The distribution of overall scores was consistently shifted to reflect knowledge improvements. These gains were similar across all four classes, suggesting consistency across all implementations of the modules. We further observed that pretest performance was predictive of overall gain: students with lower performance on the pretest showed the greatest increases in knowledge as measured on the assessment. This relationship suggests that lower-achieving students made greater gains, reducing the achievement gap between lower- and higher-performing students. For higher-performing students, gains were concentrated in the bioinformatics aspect of the test. These findings suggest that the intervention benefited students at all levels. Students who already had strong biology backgrounds tended to learn more in the relatively unfamiliar area of bioinformatics.
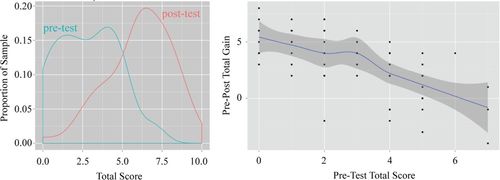
FIGURE 3. Left, density plot showing overall scores for the pre and post knowledge tests. The shift in distribution indicates increases in knowledge. Right, correlation between pre/posttest gain and initial performance on the pretest. Students who performed lower on the initial pretest demonstrated the highest gains, reflecting a narrowing of the achievement gap.
Attitudinal Survey
Students demonstrated strong attitudinal gains that were also consistent across all four classes, as shown in Table 3 and Figure 4. For the attitudinal survey results, we did not observe a correlation between low pretest values and high gain scores. The two types of gains (knowledge and attitudinal gains) were not strongly correlated; students had strong attitudinal gains regardless of how much material they learned.
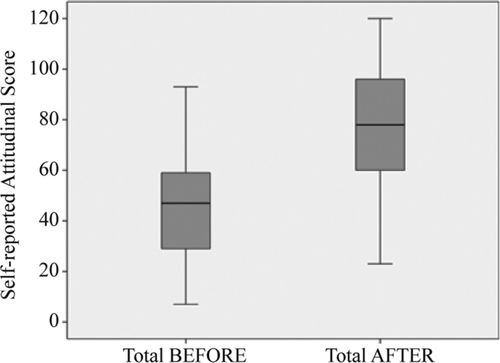
FIGURE 4. Box plot showing attitudinal gains following the Genetics of Race module. Results were based on a survey conducted after the conclusion of the module in which students self-reported their attitudes before and after the intervention.
DISCUSSION
Assessment results indicated that students who participated in the Genetics of Race experiment demonstrated both improved attitudes toward science and strong knowledge gains related to sequencing and bioinformatics. Building on the success of our initial implementations, we have continued to run this module in a range of classrooms, including standard-track biology as early as ninth grade.
We have not collected formal data on student learning regarding race and ancestry or their attitudes about race. In our informal classroom observations, students initially expect a strong link between genetics and race, whereas after the module they share the view that genetics and race are not strongly associated. We have discussed with our IRB the option of adding new assessment questions regarding attitudes about race; we hope to develop additional assessments for future implementations. We have observed that students consistently respond well to the topic of race, and teachers typically view this topic as effective in raising student engagement. This module has continued to garner interest among teachers, both those interested in rerunning the unit and those new to our project. While the topic has been a draw for many teachers, one of our local partner schools is unable to participate in this module due to a policy against racially charged topics in the classroom.
On the basis of our experience running this module, we believe that it is suitable for most high school biology classrooms. We are now working to package and distribute the Genetics of Race experiment to be run by teachers independently, as it should be possible for many well-equipped biology classrooms to implement this module without the direct support of a program such as ours. The sample-collection procedure requires common lab equipment, such as micropipettes and vortexers, as listed in the Supplemental Material. The biggest equipment challenge is the need for DNA-sequencing technology. It may be possible for classrooms to obtain access to NGS technology through coordination with local universities or community colleges or local companies that have NGS sequencers.
In addition to obtaining sequencing access, implementing this program with NGS also involves cost considerations. With an estimated class size of 25 students and two samples per student (corresponding to two hypervariable regions of the mitochondrial DNA), we process 50 samples in a typical classroom implementation. The cost of processing and sequencing these samples using the Illumina MiSeq System is approximately $2800 (roughly $2000 for consumables and $800 for technician time). This cost includes leftover consumables that can be applied toward additional students. In addition, further sequencing cost reductions are expected as the technology continues to develop. A more detailed description of costs is provided in the Supplemental Material.
While we designed this module around NGS technology to focus on the recent breakthroughs in genetics and bioinformatics, traditional Sanger sequencing can also be used as a lower-cost alternative. The use of Sanger sequencing in place of NGS would entail minor modifications to both the classroom and laboratory components of the module (Supplemental Material) and would involve shipping samples to a sequencing service rather than requiring a specialized NGS laboratory. This alternative meets the needs of the experiment, but will sacrifice the exposure to cutting-edge technology. Using Sanger sequencing, the equivalent cost for 50 samples would be approximately $1600 (estimating $1200 for consumables and $400 for technician time).
As we have designed additional classroom modules for our BioSeq program, we found that the Genetics of Race experiment functioned as an effective template for the development of these modules. Using sequencing and bioinformatics, many different topics can be addressed with similar experimental procedures and curricula. So far, we have explored diverse topics such as the human microbiome and the effects of genetic mutation on the function of microorganisms. By applying this general structure of laboratory-based modules facilitated by sequencing and bioinformatics, it is possible to address a broad range of engaging research questions to spark student interest in genetics.
CONCLUSION
The BioSeq program aims to introduce NGS to high school students while improving interest and attitudes toward sequencing and bioinformatics. To achieve these goals, we have developed laboratory-based curricular units to engage students with a personally relevant topic while also helping students become more adept with scientific protocols and more confident in their research abilities. As reported here, the initial implementations of the Genetics of Race experiment serve as a proof-of-concept for this approach. In this module, students explored the concepts of genetics and race in the context of a science classroom. Students expanded their understanding of modern methods of genetic sequencing, bioinformatics, and biotechnology research and how these methods can be applied to our daily experiences. They also practiced analyzing data with a critical mind, preparing them to better interpret and assess information. Our assessment outcomes indicate an increased interest and comfort with science for a broad spectrum of students. We continue to develop additional NGS experiments for high school students, including modules based on investigating microbiomes and studying the effects of mutations on bacteria. In the future, we plan to provide a wide variety of interesting modules and accessible resources to bring high school students closer to current research and modern genetics techniques.
ACKNOWLEDGMENTS
This project was supported by the Office of the Director, NIH, under award number R25OD010547-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.



