GenBio-MAPS: A Programmatic Assessment to Measure Student Understanding of Vision and Change Core Concepts across General Biology Programs
Abstract
The Vision and Change report provides a nationally agreed upon framework of core concepts that undergraduate biology students should master by graduation. While identifying these concepts was an important first step, departments also need ways to measure the extent to which students understand these concepts. Here, we present the General Biology–Measuring Achievement and Progression in Science (GenBio-MAPS) assessment as a tool to measure student understanding of the core concepts at key time points in a biology degree program. Data from more than 5000 students at 20 institutions reveal that this instrument distinguishes students at different stages of the curriculum, with an upward trend of increased performance at later time points. Despite this trend, we identify several concepts that advanced students find challenging. Linear mixed-effects models reveal that gender, race/ethnicity, English-language status, and first-generation status predict overall performance and that different institutions show distinct performance profiles across time points. GenBio-MAPS represents the first programmatic assessment for general biology programs that spans the breadth of biology and aligns with the Vision and Change core concepts. This instrument provides a needed tool to help departments monitor student learning and guide curricular transformation centered on the teaching of core concepts.
INTRODUCTION
The Vision and Change national report outlined five core concepts that all biology majors should master by graduation, namely 1) evolution; 2) structure and function; 3) information flow, exchange, and storage; 4) pathways and transformations of energy and matter; and (5) systems (American Association for the Advancement of Science [AAAS], 2011). Identified from conversations among more than 500 biologists and biology educators across the country, these core concepts represent a consensus view of the central ideas in biology. Furthermore, these core concepts are similar to the central biology concepts contained in the Advanced Placement (AP) Biology Curriculum Framework (Wood, 2009; College Board, 2011) and Next Generation Science Standards (NGSS Lead States, 2013), lending further credence to the community’s support for the importance of these core concepts.
Vision and Change provided an overarching framework with its broad descriptions of the core concepts and established a starting point for others to unpack these big ideas with more detail. To further articulate the core concepts, an iterative, grassroots approach incorporating feedback from more than 240 biologists and biology educators led to the creation of the BioCore Guide (Brownell et al., 2014). This framework delineates key principles and concepts underlying each core concept within three biology subdisciplines approximating the diversity of biology (i.e., molecular/cellular, physiology, and ecology/evolution), giving departments a tool to help them align their instruction with the Vision and Change core concepts.
The emergence of these overarching conceptual frameworks has led to the need for departments to have tools to assess how well they are teaching the core concepts of Vision and Change. Rubrics developed by the Partnership for Undergraduate Life Science Education (PULSE) community can be used to self-evaluate the extent to which the courses in an undergraduate program focus on the core concepts (Aguirre et al., 2013; Brancaccio-Taras et al., 2016). Other assessment tools have been developed that are aligned with the core concepts, such as the biology card sorting task (Smith et al., 2013), but these assessments cannot be practically administered to hundreds of students in a program due to the tools’ open-ended format. Existing concept inventories that are closed-ended typically focus on individual topics or courses (e.g., Smith et al., 2008; Shi et al., 2010; Kalas et al., 2013; Kalinowski et al., 2016) but do not span the breadth of topics covered in an undergraduate biology program and are not explicitly aligned with the core concepts.
By gauging student understanding across an entire major, programmatic assessment represents an important mechanism to help monitor and guide departmental progress toward achieving the goals of Vision and Change. The decision to use programmatic assessment can stimulate conversations within a department on what it intends to teach in its programs, which courses address these important concepts, and whether potential thematic linkages exist across courses (Marbach-Ad et al., 2007). Programmatic assessment data can help departments determine the extent to which students have learned various concepts at different points in a program, identify challenging concepts for which alternative teaching strategies can be employed, determine whether specific demographic characteristics relate to student performance, and monitor the impact of instructional changes (Marbach-Ad et al., 2010). Furthermore, as administrators, accreditation bodies, and government agencies call for evidence of the “value added” by an undergraduate education, programmatic assessment can provide an empirical basis for evaluating learning outcomes and justifying subsequent curricular decisions (Shavelson, 2010; Arum and Roksa, 2011; Arum et al., 2016).
Despite the potential benefits of programmatic assessment, we still lack sufficient means to directly measure at scale the extent to which students have mastered the core concepts as they advance through general biology degree programs found at the vast majority of undergraduate institutions (Brownell et al., 2014). Here, we describe the development of the General Biology–Measuring Achievement and Progression in Science (GenBio-MAPS) instrument as a tool to measure student understanding of the Vision and Change core concepts at key time points during an undergraduate general biology program. We aligned the content of this instrument to the BioCore Guide consensus framework to reflect the breadth of concepts and subdisciplinary areas covered in general biology programs. We designed GenBio-MAPS for administration at three time points during an undergraduate degree: 1) at the beginning of an introductory biology series, 2) after completion of the introductory biology series, and 3) at an advanced time point before graduation from a bachelor’s program (Figure 1). These time points enable 2- and 4-year institutions to assess students’ incoming knowledge, measure the impact of introductory courses, and determine the cumulative learning outcomes of their biology curricula. GenBio-MAPS complements the other program-level instruments developed by our group for specific biology subdisciplines, including the Molecular Biology Capstone Assessment (MBCA) (Couch et al., 2015), Phys-MAPS (Semsar et al., 2019), and EcoEvo-MAPS (Summers et al., 2018). Together, this suite of instruments provides departments with tailored ways to gauge student conceptual understanding at key junctures and inspire curricular changes to improve their programs.
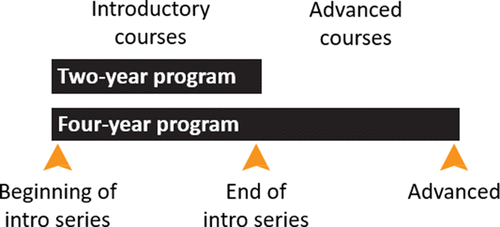
FIGURE 1. Administration time points for 2- and 4-year biology programs. GenBio-MAPS was designed to be administered at the beginning of the introductory series, end of the introductory series, and toward the end of advanced course work.
METHODS
Question Format, Development, and Revision
We used a multiple-true-false (MTF) format in which each question consists of a stem that introduces a biological scenario followed by a series of independent true–false (T-F) items (Frisbie, 1992). This format has several advantages that make it particularly suitable for programmatic assessment. First, the closed-ended nature of these questions enables rapid and consistent scoring. Second, the T-F items can probe student understanding of different concepts related to the same scenario, and students can answer several T-F items in the same amount of time that it takes to answer one multiple-choice (MC) question, enabling the test to cover a broader range of content in a limited time span (Frisbie and Sweeney, 1982; Kreiter and Frisbie, 1989). Third, the traditional MC format only captures a student’s preferred answer and thus cannot detect instances in which students have incomplete or mixed conceptions in which they believe more than one response option to be correct (Parker et al., 2012). The MTF format overcomes this issue by having students separately evaluate each T-F item, thereby providing a more detailed portrait of student thinking (Couch et al., 2018). Finally, MTF questions and other multiple-response formats have been shown to approximate the reasoning expressed by students in free-response answers and reveal specific incorrect conceptions that go underdetected in open-ended formats (Wilcox and Pollock, 2014; Hubbard et al., 2017).
In developing questions, we followed a set of guidelines to ensure consistency in style and content across the instrument. Each MTF question consists of an introductory stem followed by four to five T-F items. The question stems span a range of biological scales from molecules to ecosystems and often include a diagram, graph, or table that students must interpret. The T-F items were developed to align with the core concepts and statements specified in the BioCore Guide within three major biology subdisciplines: molecular/cellular biology, physiology, and ecology/evolution. We sought to maximize the extent to which students were required to think across the core concepts by having each stem include T-F items that addressed at least two different core concepts. This strategy also allowed us to test transfer of each core concept to a variety of contexts so that the diagnosis of student understanding of a core concept would not be solely dependent on any specific scenario. To generate questions that targeted conceptual understanding rather than factual memorization, we limited the use of scientific jargon and avoided common textbook examples. We avoided words that could provide answer cues (e.g., “never,” “always”) and maintained a relatively even balance of the number of true and false items across the instrument to prevent students from employing test strategies (Frey et al., 2005).
We developed questions using an iterative process (Figure 2) intended to optimize instrument validity and reliability (Adams and Wieman, 2011). During the first phase of question development, seven authors (B.A.C., C.D.W., S.F., J.K.K., M.K.S., A.J.C., S.E.B.) with a range of subdisciplinary expertise drafted an initial set of MTF questions, and each question writer assigned his or her T-F items to a core concept and subdiscipline. We reviewed these alignments to determine which areas needed additional coverage and identify questions that only addressed one core concept. We then added additional questions and items to help balance representation of the core concepts and subdisciplines across the question set. We conducted think-aloud interviews with 29 students at one research-intensive university to identify issues with question clarity and determine whether student answers were consistent with their underlying thinking (Anders and Simon, 1980), making iterative revisions throughout this process. An initial set of 16 questions with 73 items was piloted to 881 students in seven course sections at three institutions during Spring 2014.
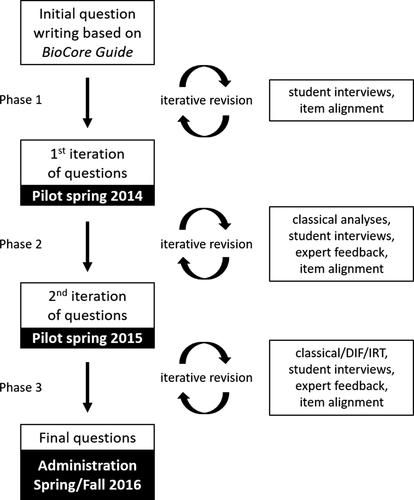
FIGURE 2. GenBio-MAPS question-development process. Assessment questions were drafted and iteratively revised over the course of three phases, each culminating in a large-scale administration. See the Methods section for further details. IRT, item response theory.
During the second phase, we analyzed results from the previous pilot using classical test theory statistics (Crocker and Algina, 2006), wrote 24 new questions, and made iterative revisions based on these analyses as well as 135 additional student interviews at one community college and four research-intensive universities spanning the country (i.e., Northwest, Southwest, Mountain West, and Northeast). We solicited feedback from 20 experts with appropriate subdisciplinary backgrounds to ensure that each question’s content was clear, scientifically accurate, and appropriate for a general biology major. Questions and items were removed when they were determined to not be performing appropriately. Two authors (C.D.W., A.J.C.) independently aligned each item to the core concept and subdiscipline that it addressed and discussed any disagreements until they reached consensus. This second phase culminated during the Spring 2015 semester when we piloted a revised set of 38 questions with 194 items to 2621 students in 49 course sections at 10 institutions.
During the third phase, we began by conducting analyses of the previous pilot data, including classical item analysis, detection of differential item functioning (DIF), and development of item response theory (IRT) models. Building on these pilot results, we drafted three new questions, and a team of four authors (B.A.C., C.D.W., A.J.C., S.E.B.) reviewed the entire question bank as a group and conducted additional revisions with particular attention to items flagged during the previous analyses (e.g., items with low discrimination, bias toward particular demographic groups, or poor fit to the model), while taking into account question performance during prior think-aloud interviews. Again, questions and items with unresolvable issues were removed. We also drafted knowledge statements to delineate the understandings targeted by each item. As the questions were finalized, we conducted 31 additional student interviews at one research-intensive university and solicited feedback from 38 experts, prioritizing feedback on new and revised questions. Two authors (C.D.W., A.J.C.) again independently aligned all the items to a primary core concept (80% agreement) and subdiscipline (88% agreement) and reached consensus on any disagreements through discussion.
The final instrument consists of 39 question stems and 175 accompanying T-F items, including 39 items on evolution; 31 items on structure and function; 41 items on information flow, exchange, and storage; 37 items on pathways and transformations of matter and energy; and 27 items on systems. These same items can also be categorized according to the subdisciplines, with 86 in molecular/cellular biology, 42 in physiology, and 47 in ecology/evolution. The full assessment and associated knowledge statements can be found in Supplemental Material 1.
Final Administration
For the final administration, each student answered a random subset of 15 question stems and associated T-F statements from the full question bank (i.e., each student answered a total of 60–75 T-F items). In addition, the order of T-F statements within each question stem was randomized for each student to minimize any item-order effects. Students also answered a set of demographic questions at the end of the survey (Supplemental Material 2). The survey as a whole was designed to take ∼30 minutes to complete.
We administered the final version of the instrument to students in 152 courses at 20 institutions with general biology programs during the 2016 calendar year (Table 1), including 11 institutions with courses at all three time points in the undergraduate major (Supplemental Material 3). We employed a cross-sectional design, meaning that different students completed the instrument at the different time points. We collected data at the first time point at the beginning of the first introductory biology course; the second time point at the end of the last course in a program’s introductory biology series, typically the second (for semester systems) or third (for quarter systems) course in the major; and the last time point at the end of upper-division courses that tended to be taken near the end of a program.
| Institution characteristic | n | % |
|---|---|---|
| Control | ||
| Public | 15 | 75 |
| Private | 5 | 25 |
| Regiona | ||
| Mid-Atlantic | 2 | 10 |
| Midwest | 10 | 50 |
| Northwest | 3 | 15 |
| Southwest | 5 | 25 |
| Carnegie basic classification | ||
| Associate’s Colleges: Mixed Transfer/Career & Technical-High Nontraditional | 2 | 10 |
| Baccalaureate Colleges: Arts & Sciences Focus | 3 | 15 |
| Master’s Colleges & Universities: Larger or Medium Programs | 7 | 35 |
| Doctoral Universities: Higher or Moderate Research Activity | 3 | 15 |
| Doctoral Universities: Highest Research Activity | 5 | 25 |
| Course time point | ||
| Beginning of introductory series | 58 | 38 |
| End of introductory series | 45 | 30 |
| Advanced | 49 | 32 |
We adopted an administration strategy that enabled us to collect and score the data in a consistent and efficient manner across institutions. Students completed the instrument in an online survey outside of class time. Each course instructor was directed to verbally announce that students, as part of normal course practices, would complete an assignment to gauge their understanding of core biology concepts. To incentivize student participation, instructors were asked to give students a small amount of regular or extra credit for the assignment, with the exact amount being at the discretion of the instructor. Students were additionally told that they would have the option to release their responses for research purposes but that this decision would have no effect on their course grade. After class, the instructor sent students a link to a Qualtrics survey. The first survey page introduced the assignment and asked students to answer the questions to the best of their abilities in one sitting on a large-format device (e.g., laptop, desktop) and avoid consulting outside resources (e.g., peers, websites). The second page of the survey contained a consent form that described the project and prompted students to indicate their willingness to release their responses for research purposes.
Data Processing, Participation Rates, and Student Demographics
We applied a stringent filtering process to generate a high-quality data set reflecting the target population. We first removed any survey submissions for which the student did not finish the survey, reported being under 18 years of age, did not consent, or had already submitted a survey in the same course. To reduce potential noise from responses containing extensive guessing, we next excluded any responses for which students completed the survey in less than 10 minutes, because this was determined to be too short a length of time to have made a good faith effort to read and answer the questions. Finally, we excluded students who did not answer at least 60 T-F items and responses from students who fell outside the target population, including students who had already taken the survey in a different course, students at the postbaccalaureate or graduate level, or students who indicated that they were not planning to major in life sciences. In total, the final data set consisted of 5175 responses, which we estimate represents 65% of the eligible students enrolled in the courses. This participation rate approximates the number of eligible students by taking overall course enrollment and subtracting an ineligible student estimate (i.e., students who were underage, enrolled in another section, postbaccalaureate or graduate status, or nonmajors) based on the ineligible response rates seen in surveys. Demographic information for students included in the final data set can be found in Table 2. The group with the most students served as the reference group for nominal demographic variables. With respect to the time points, 2425 responses (47%) came from students at the beginning of the introductory series, 1832 responses (35%) came from students at the end of the introductory series, and 918 responses (18%) were from advanced students in upper-division courses. While students enter and advance through programs at different rates, the first time point consisted primarily of first-year and sophomore students and the last time point consisted almost entirely of juniors and seniors (Supplemental Material 4).
| Student characteristic | na | % |
|---|---|---|
| Course time point | ||
| Beginning of introductory series | 2425 | 47 |
| End of introductory series | 1832 | 35 |
| Advanced | 918 | 18 |
| Class standing | ||
| First year | 2049 | 40 |
| Sophomore | 1319 | 25 |
| Junior | 1011 | 20 |
| Senior | 796 | 15 |
| Approximate current overall GPA | ||
| 4.00–3.70 (A+ to A−) | 1748 | 43 |
| 3.69–2.70 (B+ to B−) | 2896 | 56 |
| 2.69–1.70 (C+ to C−) | 362 | 7 |
| 1.69–0.00 (D+ to E/F) | 27 | <1 |
| Gender | ||
| Female | 3376 | 65 |
| Male | 1755 | 34 |
| Other | 27 | <1 |
| Ethnicityb | ||
| Non-underrepresented | 4360 | 84 |
| Underrepresented | 735 | 14 |
| English language | ||
| English spoken at home growing up | 4437 | 86 |
| English not spoken at home growing up | 722 | 14 |
| Highest parental education level | ||
| Completed bachelor’s degree | 3204 | 62 |
| Did not complete bachelor’s degree | 1886 | 36 |
| High school biology course work | ||
| No AP Biology | 3209 | 62 |
| AP Biology | 1916 | 37 |
| Transfer status | ||
| Non–transfer student | 4384 | 85 |
| Transfer student | 779 | 15 |
Statistical Analyses
We used Mplus software (v. 8) to conduct confirmatory factor analysis (CFA) with weighted least-squares means and variance-adjusted estimation to account for the categorical nature of the item responses (Brown, 2015). We used Winsteps software (v. 3.91.0) to generate Rasch models of the item responses, calculate person reliabilities, determine item fits, and conduct DIF analysis using the Mantel-Haenszel test (Linacre, 2014a). We also used the same Rasch models to generate estimates of overall student ability (i.e., theta) and modeled item difficulties in units of logits. The Rasch model estimates the probability of a student answering a particular item correctly based on student ability and item difficulty (Bond and Fox, 2007).
We used classical test theory to calculate overall student scores, core concept scores, subdiscipline scores, and item difficulties. Overall, core concept, and subdiscipline scores were calculated as each student’s percent correct across all the T-F items in that group. Item difficulty was calculated as the percent of students answering each item correctly. We compared Rasch and classical student and item metrics using Pearson correlations.
We calculated linear mixed-effects models with restricted maximum-likelihood estimation to understand how different variables explained student performance. Predictor variables were included based on whether they were hypothesized a priori to explain variance in the outcome variable: no further model selection or model averaging was performed. For the base model predicting overall scores, we included institution and course nested within institution as random effects (to account for potential differences between data-collection sites) and student self-reported demographic variables as fixed effects. 
For the two models predicting subcategory (i.e., core concept and subdiscipline) scores, we included institution, course nested within institution, and student nested within course and institution as random effects (to account for data-collection sites and repeated measures across the subcategories) and time point, subcategory, and time point × subcategory as fixed effects. 
Item differences between time points were determined by calculating the normalized difference for each item across the entire sample from the beginning of the introductory series to the advanced time point, according to the formula 
This work was approved under protocols at Arizona State University (00001058, 00003057), University of Colorado–Boulder (15-0283), University of Maine–Orono (2015-06-07), University of Nebraska–Lincoln (14618), University of Washington–Seattle (00000672), and all piloting institutions.
RESULTS
Test and Item Characteristics
In developing GenBio-MAPS questions, we wrote items that aligned with the five core concepts and three subdisciplines delineated in the BioCore Guide. We determined the extent to which these alignments could explain variation in student responses. We found that a CFA model wherein all of the questions were considered as one factor (root mean square error of approximation [RMSEA] = 0.007, confirmatory fit index [CFI] = 0.87, Tucker-Lewis index [TLI] = 0.86) yielded fit statistics similar to models that included either the five core concepts as separate factors or the three subdisciplines as separate factors (RMSEA = 0.007, CFI = 0.87, TLI = 0.87 for both models). We also found that core concept or subdiscipline factor scores were highly correlated with each other (r > 0.96 for all pairwise correlations across core concepts or subdisciplines), indicating that students exhibit similar relative performance across these subcategories and that the subcategory groupings provide little explanatory power beyond the unidimensional model.
We generated Rasch models to determine the extent to which student responses to individual items were consistent with their broader performance on the test. We analyzed person reliability as a metric for the consistency of student responses across all the items on a test. We first developed a model in which all the items were considered as a single scale, which produced an acceptable reliability of 0.82 (Kline, 2000). We also analyzed each core concept and subdiscipline as separate models and found that the reliabilities for these models were variable, ranging from 0.18 to 0.50 for the core concepts and from 0.41 to 0.72 for the subdisciplines (Supplemental Material 5). These lower reliabilities likely stemmed from the comparatively smaller number of items in each subcategory and suggest that individual student scores for core concepts and subdisciplines should be interpreted with caution. However, these scores may still be useful when aggregated at the cohort level for identifying broader performance trends.
We next sought to determine how well the individual items aligned with a student’s overall performance (Supplemental Material 6). Rasch point measures represent the correlations (point-biserial coefficient) between item responses and modeled student ability scores (Linacre, 2014b). The vast majority (172 out of 175) of the items had positive values, whereas only three items (15b, 36d, and 45d) had negative point measures, indicating that higher-performing students did slightly worse than their lower-performing counterparts. We elected to leave these three items on the instrument, because they were interpreted appropriately during student interviews, they tested important concepts, their low correlations could be explained by poor student performance, they did not hinder the overall instrument from achieving acceptable reliability levels, and they had negligible effects on total scores. We analyzed Rasch outfit mean-square statistics as a metric for the degree to which responses to each item fit the test model. For the outfit mean-square statistic, all of the items had acceptable fits based on having values between 0.5 and 1.5 (Linacre, 2014b).
We further wanted to determine whether any of the items displayed potential signs of bias based on student demographic characteristics (Martinková et al., 2017). The Mantel-Haenszel test analyzes whether two groups show significant differences on individual items beyond what would be expected given the overall scores of these students (Crocker and Algina, 2006). In analyzing the results from this test, we paid particular attention to any items with significant differences between the reference and nonreference groups that would be classified as category C according to Educational Testing Services criteria (Zwick et al., 1999; Linacre, 2014b). Category C items have moderate to large differences in the modeled difficulty for the two groups (DIF contrast ≥ 0.64). Two items (31b and 45d) met this criterion for gender, and two other items (22a and 38c) met this criterion for race/ethnicity. In both cases, one item was easier for the nonreference group, and the other item was harder for the nonreference group. We elected to leave these items on the instrument, because they showed no explicit signs of bias during student interviews, they seemingly had no distinguishing features that related to the particular demographic variable, and they had a neutral net effect on overall scores.
Comparing Rasch and Classical Metrics
Rasch modeling estimates person and item parameters based on how students answer each item. This is particularly useful for instruments such as GenBio-MAPS that use a test administration design in which students only answer a subset of all the questions, because student ability scores account for the difficulty of the particular items answered by each student. However, we also recognize that many institutions might lack the necessary expertise, software, and sample size to analyze test data using item response models. Thus, we compared Rasch analyses with classical student and item metrics to determine whether there were functional differences between these two analytic approaches. We found that Rasch student ability scores were highly correlated with overall percent correct (r = 0.97; Supplemental Material 7A). In visualizing this relationship, the vast majority of students fell along the linear portion of the sigmoidal curve, while the highest-performing students, constituting roughly 1% of the sample, fell in the upper portion of the curve. We also found that Rasch item difficulties and item percent correct values had a strong correlation (r = −0.99), with only a few of the easiest items showing deflection from a one-to-one relationship (Supplemental Material 7B). Given that most institutions using GenBio-MAPS will employ classical test statistics and that these metrics correlate very closely with Rasch-based measures, the remaining analyses will use classical test results. This data presentation strategy has been adopted previously to help make test results more interpretable for the target audience (Vincent-Ruz and Schunn, 2017; Summers et al., 2018).
Overall Student Performance, Demographic Effects, and Institutional Patterns
We next sought to understand broad student performance patterns based on overall test scores. Across institutions, students had an overall score median of 61% at the beginning of the introductory series, 68% at the end of the introductory series, and 75% at the end of advanced courses (Figure 3A). We generated a linear mixed-effects model to control for sampling variance and estimate the contributions that various factors make to overall scores (Table 3). We found that administration time point had a large impact on student scores, modeled as a difference of 6.5% from the beginning to end of the introductory series and 11.7% from the beginning of the introductory series to the advanced time point. By comparison, class standing (i.e., first-year, sophomore, junior, senior) had a much smaller effect of less than 1% change between levels. Self-reported grade point average (GPA) had an effect of roughly 3.5% change for each higher letter grade. In comparison with their reference group, we found a positive effect for students who took AP Biology in high school (2.7%). Students who were female, were from an underrepresented minority (URM) group, did not speak English at home, or did not have a parent who graduated from college experienced a negative effect attributable to these variables (−3.0, −2.0, −3.2, and −2.0, respectively), whereas we detected no significant effect for transfer students. To further investigate the effects of these demographic characteristics, we generated a priori planned models testing for potential interactions between time point and gender, ethnicity, language, or parents’ education. In each of these separate models, we found no significant effect for the interaction term, indicating that the discrepancies seen for each demographic variable remain consistent across the major and do not narrow or widen at later time points.
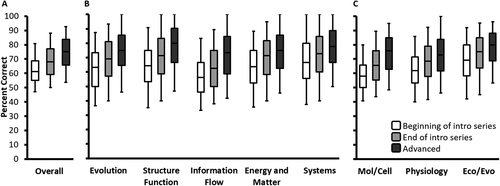
FIGURE 3. Student raw score distributions at the different time points based on (A) overall scores, (B) core concept scores, and (C) subdiscipline scores. Central bars represent median overall percent correct, boxes represent inner quartiles, and whiskers represent 5th and 95th percentiles. Post hoc Tukey’s tests revealed significant differences between all adjacent time points. Post hoc Tukey’s tests were significant between all adjacent time points, indicating that students show growth between time points.
| Parametera | Estimate | SE | df | t | p |
|---|---|---|---|---|---|
| Time point (ref: beginning of intro series) | |||||
| End of intro series | 6.53 | 0.69 | 97.2 | 9.4 | <0.001 |
| Advanced | 11.66 | 0.85 | 185.4 | 13.7 | <0.001 |
| Class standing | 0.77 | 0.23 | 3872.9 | 3.4 | 0.001 |
| GPA | 3.53 | 0.24 | 4809.3 | 14.7 | <0.001 |
| Gender (ref: female) | |||||
| Male | 3.04 | 0.29 | 4764.9 | 10.4 | <0.001 |
| Race/ethnicity (ref: non-URM) | |||||
| URM | −1.96 | 0.43 | 4783.1 | −4.5 | <0.001 |
| Language (ref: English spoken at home) | |||||
| English not spoken at home | −3.16 | 0.42 | 4769.1 | −7.5 | <0.001 |
| Parental education (ref: parent graduated college) | |||||
| No parent graduated college | −2.05 | 0.31 | 4791.4 | −6.6 | <0.001 |
| AP Biology (ref: no AP Biology) | |||||
| Took AP Biology | 2.71 | 0.30 | 4787.1 | 9.2 | <0.001 |
| Transfer (ref: non-transfer) | |||||
| Transfer student | −0.18 | 0.43 | 4811.2 | −0.4 | 0.675 |
Although we did not design the GenBio-MAPS instrument for the purpose of comparing institutions, we tested whether it has the important property of detecting institution-specific outcomes. Specifically, we added a time point × institution interaction term to the base model. This term was significant, indicating that institutions show different trajectories across the time points (Supplemental Material 8). We further plotted average raw overall student performance for the 11 institutions with data at all three points (Figure 4). These institutions showed a range of different profiles across the three time points. The patterns did not necessarily reflect different classes of institutions (based on the Carnegie basic classification), as each pattern could be observed for different institution types. In some cases, students at an institution had equivalent increases in performance between consecutive time points, suggesting continual gains across the curriculum. In other cases, students at an institution showed little difference between the first two time points, but a larger increase between the later time points or, conversely, a large difference between the first two time points followed by a smaller difference across the later time points. In these cases, a plateau between adjacent time points could highlight a time period with little growth and periods for programs to consider potential improvements.
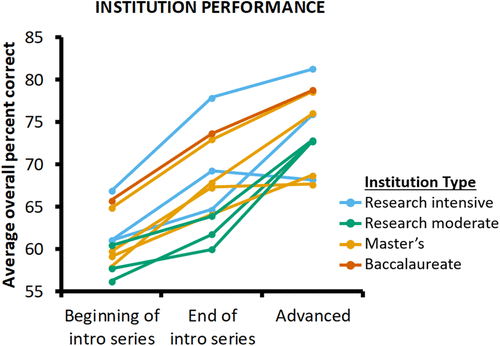
FIGURE 4. Student performance at different institutions across time points. Points represent average raw overall percent correct at the beginning of the introductory series, end of the introductory series, or advanced time points. Each colored line connects data from a single institution, and each series is colored based on institution type: blue, doctoral universities: highest research activity; green, doctoral universities: higher or moderate research activity; orange, master’s colleges and universities: larger or medium programs; red, baccalaureate colleges: arts and sciences focus.
Student Subcategory and Item Performance Levels
While overall scores can detect broader patterns in student performance, programs also need higher-resolution information to identify areas for growth. Thus, we began by plotting core concept and subdiscipline scores at the different time points (Figure 3, B and C). These scores would be expected to show similar patterns with overall scores, but they provide important information, because they reflect current or potential ways of organizing program content (Sinharay et al., 2011; Livingston, 2015). Additional mixed-effects models revealed interactions between time point and core concept or subdiscipline (core concept × time point: F(8, 20,685) = 9.73, p < 0.001; subdiscipline × time point: F(4, 10,344) = 31.19, p < 0.001). Post hoc Tukey’s tests were significant between all adjacent time points, indicating that students show growth between time points in each of the subcategories. For example, students showed improvements at each time point for the evolution core concept, with an overall improvement from a 63% median at the start of the introductory series to 75% at the end of the advanced time point.
In addition to subcategory scores, institutions can further examine performance at the item level to pinpoint specific areas of proficiency and deficiency. We identified items showing the highest and lowest normalized differences from the beginning of the introductory series to the advanced level across all institutions (see Tables 4 and 5 for the content of each item). The 10 items showing the highest differences had normalized differences above 0.6 (Table 4). The initial percent correct on these items showed a broad distribution with values scattered from 55% to 90%. In all cases, the percent correct was high among advanced students, ranging from 86% to 97%. These items spanned all five core concepts, but were mostly at the molecular/cellular level. We also identified the 10 items for which students demonstrated the lowest differences (Table 5). These items could show low differences because they were either challenging at both time points or relatively easy at both time points. For most of the items, the initial percent correct started and remained low (i.e., below 60%). Thus, these items were difficult at all levels rather than being too easy or “topped out” at the introductory level. The items spanned all five core concepts and covered a more even range of biological scales. While these items represented key conceptual areas, they often required students to apply these concepts in more complicated scenarios and may reflect “sticky” misconceptions that persist despite instruction (Smith and Knight, 2012).
| Percent correct | |||||
|---|---|---|---|---|---|
| Item | CC-SDa | Beginning of intro series | Advanced | Normalized difference | Knowledge statement |
| 14b | EV-E | 87 | 97 | 0.77 | Branch points represent common ancestors, but these ancestors are not the same as the descendant groups, which have evolved into something different. |
| 03d | SF-M | 86 | 95 | 0.63 | Mutations can confer viral drug resistance by changing the ability of a drug to bind its viral target. |
| 12d | SF-M | 67 | 88 | 0.63 | Phosphorylation activates proteins by causing a structural change that alters their biochemical properties. |
| 40c | SF-M | 75 | 92 | 0.68 | The frequency and duration of binding between two molecules depends on their biochemical properties. |
| 44d | SF-M | 69 | 91 | 0.72 | Binding of a ligand to an allosteric regulatory site induces a change in the structure and activity of the active site. |
| 04d | IF-M | 77 | 92 | 0.65 | Different transcription factor proteins are selectively expressed in different cell types, contributing to differences in gene expression between these cell types. |
| 22a | IF-P | 73 | 91 | 0.66 | Physiological process are often initiated by the production of a specific signaling molecule in response to a stimulus. A signaling molecule that is exogenously added to an organism can still elicit a downstream response in the absence of the corresponding stimulus, provided the signaling molecule can reach its intended location. |
| 27b | EM-M | 70 | 89 | 0.63 | Decreasing the area in which a molecule diffuses will increase its effective concentration and the likelihood that it will encounter its receptor. For signals that are released from a particular source, shortening the distance from the source to the receptor will result in increased probability of binding to its receptor. |
| 31c | EM-P | 90 | 97 | 0.68 | The reactions of cellular respiration are not 100% efficient, and some of the energy stored in glucose is ultimately released as heat during chemical reactions. |
| 12a | SY-M | 55 | 86 | 0.69 | Most genes are regulated by a complex array of signaling pathways. |
| Percent correct | |||||
|---|---|---|---|---|---|
| Item | CC-SDa | Beginning of intro series | Advanced | Normalized difference | Knowledge statement |
| 02d | EV-P | 58 | 59 | 0.02 | Mutations can increase the fitness of an organism. |
| 08d | EV-E | 86 | 85 | −0.08 | A pathogen can have different effects in different subgroups of a species due to underlying genetic differences between the subgroups. Genetic differences in subgroups can also drive the divergent evolution of a pathogen. |
| 15b | EV-E | 24 | 22 | −0.02 | Allele frequencies within a population fluctuate over time due to genetic drift, which is particularly pronounced in smaller populations. |
| 45c | SF-P | 45 | 47 | 0.04 | For two structures with the same volume, an irregularly shaped structure will have a greater surface area than a structure that is closer to spherical. Thus, for two structures with the same surface area, an irregularly shaped structure will have less volume than a structure that is closer to spherical. Structures that are closer to spherical provide the greatest amount of volume for a given surface area. |
| 36d | IF-M | 22 | 24 | 0.03 | Many genes involved in the formation of sex organs are located on autosomes. |
| 61a | IF-P | 70 | 67 | −0.09 | Hormones are able to circulate throughout the body and permeate into target tissues. |
| 12e | EM-M | 56 | 54 | −0.05 | Binding between two macromolecules is a reversible interaction whose frequency and duration is determined by the biochemical properties of the macromolecules and local environmental conditions. |
| 33d | EM-M | 53 | 54 | 0.01 | Small, nonpolar molecules, such as hormones, can readily cross through plasma membranes. |
| 45a | EM-P | 28 | 30 | 0.03 | Evapotranspiration from leaves draws water from the roots toward the leaves of a plant. This process does not require the plant to expend energy. |
| 32b | SY-P | 26 | 30 | 0.05 | Cellular receptors are normally either located within a cell or embedded in a cell membrane. Receptors circulating in the blood will not readily cross or become inserted into a membrane. Circulating receptors may bind to a signal but will not transduce the signal into a cellular response. |
DISCUSSION
In articulating the core concepts, Vision and Change created a conceptual framework for departments to place at the center of their undergraduate curricula. Building on these efforts, the PULSE community and others have published program-level rubrics that enable departments to self-assess their status in teaching the core concepts (Aguirre et al., 2013; Brancaccio-Taras et al., 2016; Cary and Branchaw, 2017). However, despite these important advances, the biology education community has lacked mechanisms to directly measure whether general biology programs are successfully teaching the core concepts.
In light of this need, we developed the GenBio-MAPS programmatic assessment instrument to test student understanding of the Vision and Change core concepts across the broad discipline of biology. To our knowledge, GenBio-MAPS represents the first freely available instrument designed for programmatic assessment of a general biology major. Several distinguishing features make this instrument amenable for a wide range of departments interested in gauging student understanding of the core concepts and monitoring the impact of curricular innovation. Importantly, the content of the instrument spans an entire program, and thus provides information at the program—not individual course—level, which should help departments think more broadly about the cumulative effects of their instruction, rather than evaluate individual courses. The instrument directly aligns with the detailed articulations of the core concepts in the BioCore Guide. To facilitate sampling of student thinking across the broad domain of biology, each student answers only a random subset of questions. The MTF question structure enables each core concept to be tested in scenarios ranging from the molecular to ecosystem levels, thereby measuring the extent to which conceptual understanding transfers across different contexts. Further, the T-F items target concepts at different levels of the curriculum, allowing the test to differentiate incoming from advanced students, and our results indicate significant differences in performance across time points. Finally, the closed-ended question format can be administered online and automatically scored, ensuring that survey administration can be conducted by any size department and that results can be quickly analyzed to inform curricular decisions.
Evidence of GenBio-MAPS Validity
Messick’s framework provides a useful lens for evaluating the validity of the GenBio-MAPS instrument (Messick, 1994). This framework represents a comprehensive and unified model that considers the origin, meaning, and use of student scores with respect to 1) content validity, 2) substantive validity, 3) structural validity, 4) generalizability, and 5) external validity.
Content validity in this case refers to the scientific accuracy of the questions and the extent to which the items represent the full range of biology. We largely addressed the scientific accuracy of the questions by soliciting feedback from biology experts, and the coverage of the core concepts stems from alignment of the instrument with the BioCore Guide. While the breadth of biology cannot be fully captured in any instrument, the BioCore Guide represents a thorough articulation by more than 240 biology faculty of the central ideas underlying each core concept. This framework served as a guide for our initial question drafting, and we made concerted efforts throughout the process to augment areas of limited coverage. In the final version, we had relatively even coverage of each core concept, with slightly fewer items in the systems subcategory. The integrative nature of the systems core concept made it challenging to capture in the MTF format, in which each item focuses on a specific idea, and this challenge has been reported previously with other closed-ended formats (Smith et al., 2013).
While the GenBio-MAPS questions cover a wide range of topics, we could not achieve complete coverage of all the areas within biology. With respect to the context of each question stem, there is an overrepresentation of items in the molecular/cellular subdiscipline relative to the physiology and ecology/evolution subdisciplines. The relative number of items in these subcategories mirrors the proportion of students expressing primary interest in these subdisciplines as well as the common division of an introductory biology series into a molecular/cellular semester and an organismal semester that covers physiology and ecology/evolution. Certain critical areas of biology (e.g., immunology, neuroscience, animal behavior, bioinformatics) do not have extensive representation due to their content being more specialized than expected of a general biology major. While the specific content of these courses may not be covered by GenBio-MAPS, we propose that conceptual understanding in these areas could still contribute to a student’s performance on the instrument. If these courses focus their instruction on core concepts, students may transfer knowledge to the other subdisciplines represented on the instrument.
Substantive validity captures the degree to which subjects engage in the thought processes targeted by the instrument. We addressed this form of validity by conducting nearly 200 think-aloud interviews in which students were asked to describe their thought processes behind each answer choice (Anders and Simon, 1980). These interviews captured cases in which students misinterpreted a question or used undesired strategies in selecting answers. For example, we identified questions for which students picked the right answer for the wrong reason, used superficial features of a figure to correctly answer the question, or missed the question because of misinterpretation of a word that was unrelated to the biology concept. By refining questions iteratively based on these interviews, we increased the likelihood that the selected answers accurately represented student thinking.
One challenge to substantive validity stems from the possibility that students may not put forth their best effort on a low-stakes assignment completed online, outside of class. Previous work established that these conditions produce results nearly identical to those obtained when students complete an instrument in class under similar stakes (Couch and Knight, 2015). Thus, while the conditions used in this study represent low stakes for individual students, we consider them adequate to elicit student participation and yield performances similar to what might be expected of students during class. On a related note, instructors may expect that students with little knowledge engage in purely random guessing due to the MTF format. However, evidence suggests that this perception does not align with student behaviors. Several items have percent correct values below 30% at the beginning of the introductory course series, suggesting that these items reflect student misconceptions, as opposed to random guessing. Additionally, Bayesian response models of other MTF data have revealed that a random-guessing parameter does not explain student responses (Brassil and Couch, unpublished data). Rather, when students have incomplete understandings, they still answer based on an item-specific rationale, which causes their responses to deviate from random distributions (Cronbach, 1941).
Structural validity refers to how groupings and interrelations between the different items on an instrument relate to the underlying domain. In the case of GenBio-MAPS, the five core concepts and three subdisciplines provided a priori organizational structures. CFA indicated that these structures do not provide additional explanatory power beyond a unidimensional model, suggesting that students perform similarly relative to one another across the core concepts and across the subdisciplines. Given these findings, we principally used the item alignments as a means to ensure breadth of item coverage across subdisciplines and core concepts. However, these subcategory scores may still provide useful information to departments, because they can highlight areas in which students struggle. Within the national data set, information flow proved to be the most challenging core concept across time points, which may stem from the dependence of these items on specific terminology or the ability to think across different spatial scales or ontological levels. For example, DNA has both a physical structure and information contained within its sequence of bases, and these dual natures can present challenges for students (Ferrari and Chi, 1998; Duncan and Reiser, 2007).
There are several reasons why student thinking may not divide neatly along the lines of the core concepts. First, the core concepts encompass the underlying deep features of a question, yet we do not know the extent to which students answer an item based on deep versus more superficial rationales. Indeed, experts tend to use deep question features, whereas novices tend to use these deep features to a lesser extent (Smith et al., 2013). Although think-aloud interviews allowed us to decrease the chance that students would answer an item correctly based on spurious reasons, we did not have students identify the core concept addressed by each item and thus do not know whether students answered the items in the way intended by faculty who had aligned the items with the core concepts. Second, certain biological phenomena can relate to multiple core concepts. Thus, student understanding of one core concept may overlap with understanding of another core concept for that phenomenon. For example, biological structures uniquely adapted to perform specific functions tend to arise through natural selection. Thus, the way students think about structure and function may be intimately connected to their understanding of evolutionary processes. Third, most undergraduate biology programs have not specifically aligned their curricula to the core concepts, and instructors may not be explicit about the core concepts in their teaching, so students may have trouble connecting separate phenomena that reflect the same deeper concept. For example, if an instructor is teaching about variation in the length of the loop of Henle in the kidney across species, he or she may not explicitly highlight this as an example of structure relating to function. If departments do not organize their curricula according to the core concepts or make the core concepts explicit for students, then we would not necessarily expect students to have distinct reasoning patterns for different core concepts. Further research is needed to understand whether the core concepts represent distinct domains and whether student thinking aligns more with the core concepts in programs that have transformed their curricula. Finally, with respect to the subdisciplines, the questions were intentionally written to not require highly detailed subdisciplinary knowledge, so student performance may depend more on overall conceptual understanding of biology rather than the specific subdisciplines in which they have taken the most courses.
Messick’s validity framework also considers how generalizable an instrument is beyond the immediate item set and study population. In part, generalizability considers whether performance on the given items represents student understanding of the broader construct domain or whether an alternative set of items from the same domain would have yielded different results. The generalizability of GenBio-MAPS items stems from each core concept being tested by 27–41 items situated in a variety of biological contexts spanning the entire scale of biological organization. While contextual features of questions and items (organism, direction of change, etc.) may have influenced student responses to an individual item (Nehm and Ha, 2011; Heredia et al., 2016), the distribution of concepts across multiple scenarios strengthens the instrument and capitalizes on the MTF format. Because each core concept is tested in many different contexts, a student’s performance on a core concept is not determined by his or her familiarity with a single biological context.
Generalizability also pertains to the range of students involved in the initial development efforts and the extent to which the instrument would produce similar findings in other populations. During the question-development process, we attempted to maximize the diversity of student interview subjects by recruiting students from courses at different levels at a diverse set of institutions in different geographical areas. We leveraged having a multi-institution team of researchers to interview nearly 200 students; this number greatly exceeds what has been done for previous concept inventories and commercial tests, such as the AP Biology exam. Given the large scope of the instrument, this comprehensive effort was critical to ensuring that the questions would be interpretable by a broad range of biology majors. We also conducted pilot and final administrations of GenBio-MAPS at a wide variety of institution types, including community colleges. Taken together, the scope of the development process and final analyses support the use of this instrument at most undergraduate institutions with general biology programs.
External validity considers the degree to which scores correlate with other relevant measures. We found that GenBio-MAPS scores demonstrated convergence with administration time point and GPA, and these variables had the highest estimates in the linear models. This meets the reasonable expectation that students who are more advanced in a biology series or have achieved higher grades would perform better on the instrument. As many advanced courses also had smaller class sizes compared with introductory courses, it is possible that the effect of time point could be due in part to going from larger to smaller class sizes. However, we note that the effect of class size is partially accounted for in our model by the random effect for institution, because class size is generally related to program enrollment. Interestingly, class standing (first-year, sophomore, etc.) had a much smaller effect than time point, suggesting that being in college for a longer period of time does not explain performance as much as advancement through a biology program.
Approaches to Using GenBio-MAPS to Assess and Improve a Curriculum
Despite widespread support for the curricular goals outlined in Vision and Change (AAAS, 2015), departments have had few choices for directly measuring student understanding of broad core concepts across a general biology major. As a programmatic assessment instrument aligned with these core concepts, GenBio-MAPS addresses this need and can guide formative discussions within departments on how to improve their undergraduate programs through several approaches.
GenBio-MAPS provides a wealth of information on student performance at the overall, core concept, subdiscipline, and item levels. Departments can use these results to identify areas of proficiency and deficiency throughout their programs and guide curricular changes to address problem areas. For example, instructors teaching an introductory series could identify a challenging concept to incorporate at multiple points across the course series to help students build and refine their understanding. Instructors who teach advanced courses could identify concepts that remain challenging at the end of the introductory series so that these concepts can be revisited before moving on to more complex phenomena that build on these concepts. Furthermore, this type of targeted thinking could inspire broader conversations at the department level about when and how often key concepts should be integrated across a program to ensure that students graduate with robust understandings.
As a measurement instrument, GenBio-MAPS provides a means for departments to establish baseline scores and determine the impact of curricular changes on student understanding of core concepts. Departments could administer GenBio-MAPS before and after a major effort to realign their major with the Vision and Change core concepts in hopes that their efforts will yield improved outcomes. Departments may also wish to collect assessment data to ameliorate concerns that a controversial curricular change has a negative impact. For example, performance data could help diminish apprehensions associated with transforming an introductory course series to focus more on concepts than content, replacing traditional single-topic labs with a yearlong course-based undergraduate research experience or shifting the required courses for a major. Importantly, this data-focused approach to curricular thinking overlaps with departmental requirements for institutional reporting, performance reviews, and accreditation (Beno, 2004; New England Association of Schools and Colleges, 2011).
Programmatic assessment can also be used by departments to understand how students perform based on certain demographic characteristics or participation in success programs, such as bridge experiences or learning communities (Ashley et al., 2017). We found performance differences attributed to gender, race/ethnicity, language, and parental education. These results indicate that programs need to account for these variables when analyzing group performance, because group composition may change across time points and between cohorts. Furthermore, these results highlight the need for investigation at the program and national levels into why these groups perform differently (Eddy and Hogan, 2014; Wright et al., 2016) and how programs might alter their instruction to better serve the needs of all students (National Research Council [NRC], 2011; President’s Council of Advisors on Science and Technology, 2012).
Finally, GenBio-MAPS could help facilitate transition of transfer students from 2-year programs to 4-year programs or from one 4-year institution to another 4-year institution. At the aggregate level, transfer students performed similarly to their peers. However, given that the introductory course curriculum typically differs across institutions, administering GenBio-MAPS specifically to transfer students at common transition points could help both 2- and 4-year programs identify specific areas to bolster to ensure posttransition success. This approach would be particularly informative for situations in which large numbers of students follow a relatively common pathway from one set of institutions to another (e.g., from a community college system to a university system) and could help guide conversations among institutions about curricular structures.
In administering GenBio-MAPS, large departments will likely have enough students to see statistically significant differences, while smaller departments may need to combine data over multiple years to achieve sufficient sample sizes. While each student sees only a subset of the questions, our results showing correspondence between classical scores and Rasch measures of person ability suggest that this question sampling strategy does not have a large influence on overall percent correct scores (although response modeling could address any potential concerns about a student seeing different questions across time points for longitudinal-study designs). To maximize student participation and motivation, we recommend, based on our experiences, that instructors provide students with participation credit for completing the instrument and convey to students how the survey results will be used to improve undergraduate biology instruction.
How to Obtain and Administer GenBio-MAPS
We have established an online portal (http://cperl.lassp.cornell.edu/bio-maps) where interested users can access and coordinate the administration of GenBio-MAPS and other instruments developed by our group (e.g., Molecular Biology Capstone Assessment, Phys-MAPS, EcoEvo-MAPS). This portal enables users to set up survey start and end dates, generates a unique Qualtrics link where students can take the assessment, and sends a list of participating students along with an aggregated score report after the survey has closed. Users do not need a Qualtrics license to administer through this site. Users wishing to conduct research using GenBio-MAPS should contact the corresponding author for more information on data accessibility.
ACKNOWLEDGMENTS
We thank the site coordinators who helped coordinate administration, the instructors who administered the survey in their courses, the experts who provided question feedback, and the students who participated in the research. We thank Leif Saul for generating artwork for the question figures; Peggy Brickman, Jenny McFarland, and Pamela Pape-Lindstrom for serving on our project advisory board; and Cole Walsh and Natasha Holmes for online portal development. This work was supported by a collaborative grant from the National Science Foundation Transforming Undergraduate Education in Science, Technology, Engineering and Mathematics program to the University of Colorado–Boulder (B.A.C., J.K.K.; DUE-1322364), University of Maine–Orono (M.K.S.; DUE-1322556), and University of Washington–Seattle (S.F., A.J.C., S.E.B.; DUE-1323010).