Engaging with Molecular Form to Understand Function
Cells are bustling factories with diverse and prolific arrays of molecular machinery. Remarkably, this machinery self-organizes to carry out the complex biochemical activities characteristic of life. When Watson and Crick published the structure of DNA, they noted that DNA base pairing creates a double-stranded form that provides a means of accurately copying the genetic information. Understanding this link between form and function is important for understanding the basis of any biological activity. At the most basic level, a biomolecule's function is dictated by its structure. The molecule's shape and chemical properties facilitate interactions with other molecules and determine its role in the cell. Protein function depends on the precise folding of encoded linear stretches of amino acids into three-dimensional shapes. Protein misfolding can lead to various disease states. In this review, we explore engaging online resources that highlight the connection between structure and function in biomolecules. These resources are particularly relevant for instruction at the advanced high school and undergraduate biology levels.
THE STRUCTURES OF LIFE
The “Structures of Life” section of the National Institutes of Health (NIH) National Institute of General Medical Sciences website (http://publications.nigms.nih.gov/structlife/index.html) provides an introduction to biomolecules via several resources. A short movie (5:06 min) begins by demonstrating the connection between structure and function in everyday objects and then in biomolecules (Figure 1). The movie illustrates how knowledge of a protein's structure enables researchers to identify potential drug targets. Finally, the viewer learns how x-ray crystallography and nuclear magnetic resonance spectroscopy are used to determine protein structure. Text-based materials expand on the topics addressed in the movie; these can be accessed in multiple formats: Web pages, a PDF file, an EPUB, or a free paper booklet. These materials also include vignettes of undergraduates, graduate students, and researchers who are studying different aspects of structural biology, capturing some of the diversity of individuals and research in this field. Both the movie and text-based materials focus primarily on protein structure. While they talk about structure determining function, more specific examples and illustrations would assist students in visualizing this relationship. Several such examples can be accessed via links in the “Spotlight” section of “The Structures of Life” home page.
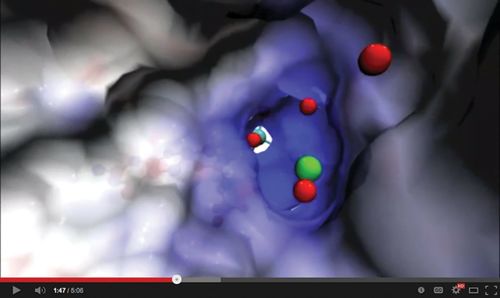
Figure 1. The Structures of Life movie illustrates how the shape of enzymes enables them to facilitate biochemical reactions. A description of the animation of the sulfite oxidase active site, shown here, is available at http://images.nigms.nih.gov/index.cfm?event=viewDetail&imageID=2746. NIH National Institute of General Medical Sciences. (http://publications.nigms.nih.gov/multimedia/video/structures-of-life.html)
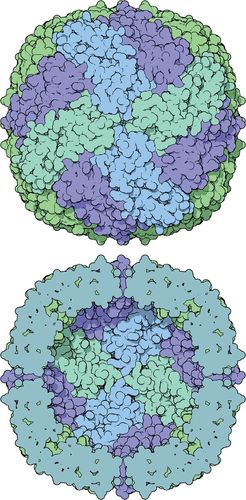
Figure 2. PDB-101's “Molecule of the Month” illustration of the iron storage protein ferritin reveals how assembled protein subunits form a hollow sphere with pores for iron internalization. Illustration by David Goodsell, the Scripps Research Institute and RCSB PDB. (www.rcsb.org/pdb/101/motm.do?momID=35)
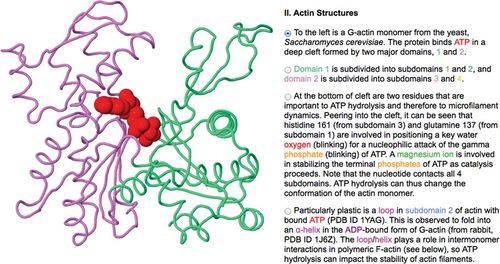
Figure 3. The Online Macromolecular Museum's exhibit on actin shows how the two major domains of an actin protein form a cleft in which ATP binds. David Marcey, California Lutheran College. (www.callutheran.edu/BioDev/omm/jmolxx/myosin_actin/myosin_actin.html)
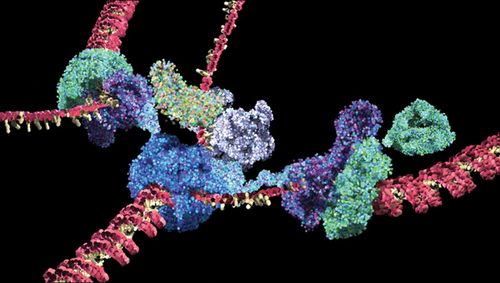
Figure 4. The HHMI and Dolan DNALC's Mechanism of Replication animation uses structural data to animate how the protein machinery unwinds, separates, and copies each strand of DNA. Copyright HHMI. (www.hhmi.org/biointeractive/dna-replication-advanced-detail and www.dnalc.org/resources/3d/04-mechanism-of-replication-advanced.html)
THE PROTEIN DATABASE (PDB)
The Research Collaboratory for Structural Bioinformatics (RCSB) maintains an international repository of three-dimensional structures for biomolecules called the Protein Database (PDB), available at www.pdb.org. Structural information in the form of three-dimensional atomic coordinate files (.pdb) is freely available for more than 130,000 biomolecules from hundreds of organisms. To make the PDB more accessible to students and educators, the PDB-101 website provides an entry point to the structural information in the database and supplies complementary educational resources (www.pdb.org/pdb/101/structural_view_of_biology.do). PDB-101 is largely organized around David S. Goodsell's visually striking “Molecule of the Month” feature, which highlights the structure and function of a molecule, usually a protein, with high-quality illustrations based on PDB structural data. For example, Goodsell's illustrations of ferritin effectively depict how protein subunits fit together to form a porous shell to uptake and store iron (Figure 2). “Molecule of the Month” addresses the importance of the featured molecule in cellular, tissue, organismal, and health contexts. Although the content is very detailed and a basic understanding of molecular biology is assumed, the language is approachable and contains minimal jargon. Because structural information about a molecule often comes from a variety of organisms and in a variety of conditions or conformations, Goodsell points users toward pertinent structural data files. Embedded links to these interactive PDB files and a short list of relevant references encourage readers to explore the structural data and literature. The “Molecule of the Month” content can also be accessed from PDB-101's “Structural View of Biology” interface, which nicely organizes the featured molecules (currently more than 80) by functional category.
PDB-101's “Educational Resources” tab contains structural posters, paper model tutorials, lesson ideas, and animations. Among the animations, What Is a Protein? (3:39 min) is a useful introduction to protein structure and the relationship between form and function. The movie introduces amino acid chemistry and the four levels of protein structure. An animation illustrates how the structure of hemoglobin enables heme binding and oxygen transport. The connection between protein form and function is also highlighted for proteins involved in immunity, nutrient storage, cytoskeleton, enzymatic activity, cellular communication, and transport. The molecular depictions are based on published structural data, and the animations effectively depict molecular interactions and conformational changes. While not integral to the accuracy of the information presented, a less repetitive soundtrack and narration by a professional voice-over artist would enhance the quality of the movie.
Finally, PDB-101's “Understanding PDB Data” section outlines how PDB structural data are generated and how these data files can be downloaded, visualized, and manipulated. This section also outlines the three major types of molecular diagrams: 1) wire frame, in which covalent bonds are depicted as lines; 2) space filling, in which each atom is represented as an electron cloud; and 3) backbone and ribbon, which illustrates secondary and tertiary structures. Each type of diagram is useful for visualizing different aspects of a molecule's structure.
INTERACTING WITH MOLECULAR MODELS
Several freely available programs are available for working with and visualizing atomic coordinate files from the PDB. One of these, PyMOL, is freely available for educational use at http://pymol.org. Jmol: An Open-source Java Viewer for Chemical Structures in 3D (www.jmol.org) is an applet designed for use in Web pages. Websites that run Jmol require that a user's computer have the latest version of the Java software (www.java.com/en) and that Java applets are enabled in its Web browser; no other software is required. FirstGlance in Jmol (http://bioinformatics.org/firstglance/fgij) is a simple platform with sophisticated functionality for viewing a molecule's structure with different diagrams, cross-sections, and emphasis on various molecular features. Currently, many sites, including the PDB, are converting from Jmol applets to the HTML5/Javascript-based JSmol, which works on tablets (e.g., iPads). The Web-based Jmol and JSmol also allow for the construction and exploration of online tutorials that guide the user through the visualization of a molecule.
An excellent source for molecular visualization tutorials is the Online Macromolecular Museum (OMM; www.callutheran.edu/Academic_Programs/Departments/BioDev/omm/exhibits.htm) maintained by David Marcey at California Lutheran College. Marcey has undergraduate students delve into the scientific literature and structural data to author molecular tutorials, which he curates and adds to OMM. The website also explains how users can build their own tutorials and includes information on working with Jmol script. OMM is organized by topic area into various “halls” (e.g., DNA/RNA, virology, immunology, signal transduction), each containing “exhibits” that step users through exploration of a molecular structure. Some tutorials use the outdated molecular visualization tool Chime, which requires its own plug-in (www.umass.edu/microbio/chime/getchime.htm#download), but these are being phased out in favor of Jmol and JSmol. OMM tutorials provide a view of the molecule on the left and the tutorial text with functional radio buttons on the right (Figure 3). Each exhibit provides useful Web browser and tutorial navigation tips. An introduction to the molecule is provided at the beginning of the tutorial, but some knowledge of biology and types of molecular diagrams is a prerequisite. The tutorial guides users with a series of radio buttons that load new structures, animate molecules, or alter the molecule's display to highlight specific features. Animations of conformational changes and binding interfaces between molecules connect details of a molecule's structure to its function. The user also can rotate the molecule by clicking and dragging to engage with the three-dimensional form. The inclusion of load/reset molecule buttons is useful here. Exhibits conclude with a list of references for further study. Because of the high level of detail, the exhibits work best at the undergraduate level, as supplements to a textbook or classroom instruction. They also lend themselves to self-guidance, so users can proceed at their own pace and replay animations. Large volumes of structural data can make the tutorials slow to load.
Other sites that include tutorials exploring molecular structures include the collaborative wiki Proteopedia (www.proteopedia.org/wiki/index.php/Main_Page), Milwaukee School of Engineering's Center for BioMolecular Modeling (http://cbm.msoe.edu), and Eric Martz's MolviZ (www.molviz.org).
ANIMATING STRUCTURE AND FUNCTION
The expanding collection of structural information has given rise to animations that capture how structural features lead to function in a cellular context. The Howard Hughes Medical Institute (HHMI) and the Dolan DNA Learning Center (DNALC) collaborated on the creation of animations (ranging in length from 39 s to 3:04 min) that harness knowledge of structural information to show molecular function. These molecular animations are available through HHMI's BioInteractive site (www.biointeractive.org/animations) and the Dolan DNALC's three-dimensional animation library (www.dnalc.org/resources/3d). Although there are multiple ways to search for the animations on the HHMI BioInteractive site, the most direct approach is to access all of their animations via the above URL and then select “DNAi” under “Series” in the left-hand column on the Web page. The URL for the Dolan DNALC website provides a direct link to the animations. Each site also contains unique content that is worth exploring.
Four of the animations (How DNA Is Packaged, Transcription, Translation, and DNA Replication) are available with both a basic and an advanced level of narration; the basic level of How DNA Is Packaged is only available on the Dolan DNALC site. For example, the Mechanism of Replication animations (Figure 4) show how DNA's double-stranded structure serves as a dual template. Both the basic and advanced versions capitalize on resolved structural information to show the proteins that facilitate DNA replication and how one strand is copied “backward” due to the antiparallel nature of the DNA strands. In the basic version (1:07 min), viewers see how molecules fit together to form the complex replication machinery, which underscores the importance of molecular structure. The advanced version (2:20 min) goes into greater detail about individual components of the replication process, adds labels, and focuses on specific features of DNA replication.
As the resources reviewed here illustrate, the field of structural biology is generating a wealth of information linking molecular form and function. Helping students build these connections can make complicated molecular phenomena more accessible.
ACKNOWLEDGMENTS
We thank A. Malcolm Campbell for critical comments and helpful suggestions on this feature.




