The Genomics Education Partnership: Successful Integration of Research into Laboratory Classes at a Diverse Group of Undergraduate Institutions
Abstract
Genomics is not only essential for students to understand biology but also provides unprecedented opportunities for undergraduate research. The goal of the Genomics Education Partnership (GEP), a collaboration between a growing number of colleges and universities around the country and the Department of Biology and Genome Center of Washington University in St. Louis, is to provide such research opportunities. Using a versatile curriculum that has been adapted to many different class settings, GEP undergraduates undertake projects to bring draft-quality genomic sequence up to high quality and/or participate in the annotation of these sequences. GEP undergraduates have improved more than 2 million bases of draft genomic sequence from several species of
INTRODUCTION
Genomics is a new and expanding field with an increasing impact on biological research and studies of human health. Genomic approaches can provide new insight to many long-standing biological questions. Instead of studying a single gene, biologists can now study entire genomes, or track genomic changes among related species. “Metagenomics” is taking this approach one step further to analyze the DNA of whole populations. Genome sequencing is constantly getting cheaper, and the “$1000 human genome” is within sight, with profound consequences for the practice of medicine (Pettersson et al., 2009). Full realization of the potential of these new developments requires a broad effort to introduce genomic approaches and bioinformatics tools into the undergraduate curriculum.
Although presenting several challenges, genomic approaches generate accessible and inexpensive research opportunities for undergraduates. The importance of providing undergraduate research experiences has been validated from several points of view. A recent report from the National Academy of Sciences, “BIO 2010: Transforming Undergraduate Education for Future Research Biologists” (National Research Council, 2003), recommends that undergraduate students learn current research methods and skills as early as possible in their education. Data indicate that a research experience gives students confidence and a sense of empowerment (BIO 2010). Bauer and Bennett (2003) report positive links between participation in undergraduate research and improved retention in science and the pursuit of graduate education. Doyle (2000) has found strong positive correlations between undergraduate research that leads to publications in refereed journals and the production of new scientists. In a 2004 study of 1135 undergraduates representing 41 universities, 91% of the subjects reported that their research experience sustained or increased their interest in postgraduate education (Lopatto, 2004). Considering the question more broadly, Locks and Gregerman (2008) have found that students who participate in research complete their science programs in greater numbers than those who do not.
The issue of retention is particularly important in overcoming minority underrepresentation in the sciences at all career levels, a major challenge for our nation. Researchers find that all students, including at-risk and first-generation minority students, benefit from undergraduate research experiences (Elgren and Hensel, 2006; Lopatto, 2006; Goins et al., 2009). Undergraduate research can influence career pathways for members of underrepresented groups by increasing the retention rate of minority undergraduates (Nagda et al., 1998) and by increasing their rate of participation in graduate education (Hathaway et al., 2002).
Colleges and universities, however, are not always able to provide independent research experiences for the majority of their students. The cost of equipment, supplies and laboratory space, and support for trained mentors on the scale needed, is beyond the budgets of many institutions. The difficulties are amplified at schools that have a high student-to-faculty ratio, who lack a pool of graduate students and postdoctoral researchers who might serve as mentors, or are generally undercapitalized. Given that many institutions share these challenges, the BIO 2010 report supports research-based laboratory courses that are designed to encourage independent or small group investigations as alternatives when individual research opportunities are limited.
We have developed the Genomics Education Partnership (GEP) to help incorporate genomics-based undergraduate research into the biology curriculum in colleges and universities across the country. GEP has grown into a partnership of diverse schools, including both primarily undergraduate institutions (PUIs) and research universities. Through the GEP project, faculty gain training and resources enabling them to introduce students to research based on genome science. Using computers and Internet access, students are given opportunities to make discoveries, learn research methods, observe the interdisciplinary nature of biological science, appreciate the importance of collaboration, and understand the connection between their classroom activities and the real world. The GEP has been designed from the beginning to allow flexibility for faculty to offer research in this field as either an independent experience or as a classroom activity, either as a stand-alone course or as part of the laboratory in a broader course in genetics/genomics/molecular biology. This flexibility has allowed the program to work within very different curricula, serving diverse students in very different institutions.
The current genomics research goals of the GEP center on an investigation into the differences between heterochromatin and euchromatin by using a comparative genomics approach. In particular, we are examining the properties and evolution of the distal portion of the dot chromosome (Muller F element) in
Here, we describe the organization and growth of the GEP, as well as our analysis of the impact GEP participation has had on both students and faculty. Although students are sometimes initially bewildered by the expectation of their making a novel contribution, most have ultimately been very enthusiastic about this approach and have reported marked personal growth as a result of participation. Faculty members also report that involvement with the GEP has been an overall positive experience, helping both them and their institutions to move ahead in genomics.
MATERIALS AND METHODS
Technical Infrastructure
The entire system is organized around a pair of SUSE Enterprise Linux servers that host a variety of services used by the GEP community. All services are available from the main website at http://gep.wustl.edu. Links from the main site allow access to information on the organization and membership of the GEP, information on workshops, curriculum and teaching materials, access to research projects, examples of prior student work, as well as assessment and communication tools. The communication tools are a bulletin board system that provides a location for timely, informal discussions, troubleshooting, and brainstorming, and a wiki-based system that houses material of a more permanent nature. The wiki system includes course syllabi devised by different GEP members, new curriculum materials produced by members, working drafts of joint manuscripts, and other work in progress. The main server also acts as a gateway to online tools that facilitate the distribution of projects to, and collection of student-generated analyses from, participating institutions. Additional tools that support the students in their research, such as a web-based program to check gene models for consistency and a web-based viewer for visualizing Basic Local Alignment Search Tool (BLAST) output, are also available.
Project Creation
All projects, whether for finishing or annotation, are compressed and uploaded to the GEP servers. Faculty members participating in the GEP can then use the Project Management System to claim and download the project packages for their students. Faculty who claim projects in a given academic year submit back their students' work each summer so that projects can be assessed and either documented as completed or (if necessary) placed back in the pipeline to be claimed during the following academic year.
To minimize costs, we rely as much as possible on publicly available data, including published draft quality genome assemblies (Clark et al., 2007) and the Trace Archive at the National Center for Biotechnology Information (see www.ncbi.nlm.nih.gov/Traces). The region of a genome to be analyzed is divided up into 40-kb projects, a work unit that we find can be handled by a single student or a small number of students working together.
Sequence Improvement Projects (“Finishing”). For any region of interest the files for the published draft sequences are consulted to ascertain the positions of fosmid clones (generally ∼40 kb) based on end sequence data. A “golden path” of overlapping clones is selected to cover the region (e.g., the dot chromosome, from the most proximal to the most distal gene), and these clones are procured from the Drosophila Genomics Resource Center. The fosmids are purified and restriction digests are prepared at the Washington University Genome Center using four different enzymes.
The student packages are created by first collecting all of the sequencing traces produced in the whole-genome project for the species and region of interest from the Trace Archive. The trace files are renamed for compatibility with the Consed suite of finishing software using the St. Louis naming standard (see Consed documentation; Gordon, 2003) and bundled together with the restriction digest data files and other support files. Each project is given a difficulty ranking based on the number and type of gaps, density of repeats, and likelihood of misassemblies, based on an initial analysis using Consed. The purified fosmid clones are retained at Washington University and used as templates for finishing reactions by using student-designed primers to obtain additional sequence data. Students are asked to finish their project to the same standard as the mouse genome; this requires 1) a complete assembly (no gaps), 2) adequate coverage of all regions (ideally, sequence from both strands), 3) resolution of ambiguous results, and 3) high quality of sequence data (Phred score of >30; The Genome Center at Washington University, 2004). As a final check, student finishers compare an in silico restriction digest of their assemblies with the results generated from the cloned DNA.
Annotation Projects. For regions of interest (either previously finished GEP sequences or high-quality draft sequences), we create overlapping projects that are between 40 and 60 kb. We use the published computational gene predictions (Clark et al., 2007) to try to avoid splitting a single gene between projects, but there are no guarantees. We analyze each project with various bioinformatic analysis algorithms (e.g., ab initio gene finders, repeat analysis, splice site predictors, conservation analysis). Using the output, we rank the difficulty of each project (on a scale of 1–3) based on the number of putative genes and the number of putative isoforms per gene. We then create a custom installation of the University of California, Santa Cruz, genome browser (Karolchik et al., 2008; see http://genome.ucsc.edu) that allows students to view the results applied to their projects (see below). Finally, the sequence of each section is bundled with all the raw data files into a package and made available on the GEP claim system. Students who are given projects are asked to derive the best gene models, including a search for all isoforms documented in D. melanogaster, by using multiple sources of information, and to report their findings to the GEP.
Surveys and Statistics
Students in GEP courses were asked to complete two sets of online instruments. As their academic term began, students completed an online 85-item survey to establish their previous experience, attitudes toward science, and learning style. The survey is based on the Classroom Undergraduate Research Experience and Summer Undergraduate Research Experience surveys (SURE); see www.grinnell.edu/academic/psychology/faculty/dl/sure&cure),with additional items that specifically target GEP course activities. Students completed a similar survey (98 items) at the end of their course in which they evaluated their learning gains on course- and research-related items, as well as giving an overall evaluation of the experience and their attitudes toward science. In addition, students were asked to voluntarily complete a test of knowledge, both pre- and postcourse. The test, designed by GEP faculty, covers basic knowledge of genes and genomes (20-question annotation quiz) and basic knowledge of sequencing reactions and data analysis (25-question finishing quiz) in a multiple-choice format.
For analysis of the test data, participating students were divided into two groups, students who had instruction in both annotation and finishing and students who had instruction in annotation only. In addition, GEP faculty recruited students at their institutions who had completed the same prerequisites but were in courses that did not use GEP material to serve as a comparison group. To encourage participation, students who completed the self-report surveys or the quizzes were given the opportunity to enter a raffle for $50 gift certificates redeemable at an online bookstore.
Course enrollment data supplied by faculty participants for 2008–2009 indicated that 472 students were enrolled in GEP-associated courses. Response rates for GEP students on the various assessment instruments were 75% for the precourse survey, 69% for the precourse quizzes, 49% for the postcourse survey, and 45% for the postcourse quizzes. In addition, 61 students not involved with the GEP courses completed the quizzes, providing a comparison group. Within the larger group of GEP respondents, 192 participants submitted complete data (all four items). These data provided the means for investigating the relationships among the measures. Approval to conduct assessment of student learning for scholarly purposes was obtained from the local Institutional Review Board (IRB) at each participating institution.
GEP faculty partners completed a survey with 25 items to indicate which ones they emphasized in their courses. In addition, faculty provided enrollment information, estimates of time on task for annotation and finishing and demographic information for their school. In late spring 2009, participating faculty completed an additional survey in which they evaluated their own experience with the GEP (Washington University IRB approval). In addition, teaching assistants (TAs) working with the GEP faculty supplied postcourse observations of the course and student learning during the first year of implementation. During the 2008–2009 academic year, 46 institutions have had students participate in the GEP research-based activities (16 partner schools did finishing and annotation, 30 did annotation only). Overall, 47 schools provided institutional information, 26 institutions provided precourse student survey/quiz data, and 22 institutions provided postcourse student survey/quiz data.
RESULTS
Structure and Organization of the GEP
The GEP arose from the successful implementation of a genomics laboratory course at Washington University in St. Louis (Research Explorations in Genomics, Bio 4342), a collaborative effort by members of the Genome Center, the Department of Computer Science, and the Department of Biology (S. Elgin, C. Shaffer, J. Buhler, E. Mardis). We had observed many didactic benefits (discussed below) of taking large research projects and breaking them down into smaller “student-sized” work units that would allow each student to tackle his/her own individual research problem, while contributing to a larger analysis. Initial studies, focusing on a comparative analysis of D. melanogaster and D. virilis dot chromosomes, have been published (Slawson et al., 2006); a second manuscript has been submitted (W. Leung, C. Shaffer, T. Cordonnier, J. Wong, M. Itano, E. Slawson Tempel, E. Kellman, D. Desruisseau, C. Cain, R. Carrasquillo, personal communication).
This experience indicated that genomics is an excellent area for introducing students to research thinking in the academic year classroom. Advantages of in silico research include the following:
requires only a computer with Internet access; therefore, widely accessible, as wet bench lab space (often in short supply) is not required;
lends itself to student/scientist partnerships—many small contributions can lead to interesting results;
lends itself to peer instruction;
has no major lab safety issues;
is effective with a short time line—computational “experiments” take minutes to run, allowing errors to be quickly recognized and experiments to be redesigned and rerun more quickly than possible with most wet bench investigations; and
is practical for a larger number of students than typically can be handled in a research lab; all students can be taught a common set of problem-solving techniques that they can then apply to their own particular project.
The initial support and enthusiasm for a nationwide project came about through a workshop at Washington University where the system was demonstrated and the potential benefits and outcomes of research-based curriculum centered on computer-based genomic analysis were discussed with a group of PUI faculty. Based on the recommendations from that meeting, appropriate support features were designed, and an initial grant proposal was subsequently submitted to and funded by the Howard Hughes Medical Institute (HHMI) Professors Program to establish the group and provide the needed central sequencing, computer infrastructure, and technical support.
Several core concepts guided the initial stages of the growth and development of the GEP. These are based on principles of inclusion and flexibility. Our goals are to create materials to teach the needed software and strategies to undergraduates, train faculty and teaching assistants at workshops in the use of these materials, and support GEP members as they incorporate this research-based curriculum into their classes, giving their students the opportunity to join in an ongoing research project.
The GEP has grown by ∼15 schools each year for the past 4 yr. Faculty join by attending a week-long workshop to learn the software and bioinformatics tools in use, as well as general strategies for implementation. All faculty are invited to send one student for similar training; these students then act as TAs to assist with initial implementation. Training workshops are scheduled in June for faculty, in August (just before the fall semester) for both faculty and TAs, and in January (just before spring semester) for TAs. There are currently 65 members in the partnership (Figure 1); approximately 48 members contributed data to the analysis reported here (members joining in 2006, 2007, and 2008).
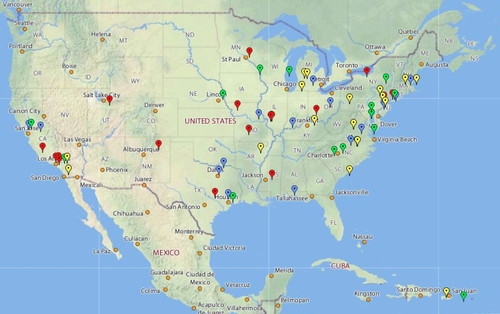
Figure 1. Members of the GEP are located on a map of the United States, color coded by the year they joined. Red, joined in 2006; blue, joined in 2007; green, joined in 2008; and yellow, joined in 2009. For current membership, see http://gep.wustl.edu.
To obtain a better understanding of the variety of institutions in the GEP, we conducted a voluntary institutional survey of the 28 private and 20 public institutions that had joined the GEP through 2008. A summary of the information from the 47 schools that responded demonstrated the diversity of the GEP member institutions. For example, an analysis of the students at GEP institutions reveals:
13 schools have <2000 students, whereas 10 schools have >10,000 students;
11 schools have >80% of students living on campus, whereas in 12 schools >90% commute;
13 schools have <5% nontraditional (>25-yr-old) students, whereas four schools have >40% nontraditional students;
six schools have >40% first-generation students; and
13 schools have >40% minority students.
creating a new, semester-long upper-level lab course based on GEP projects;
integrating a GEP project into a broader molecular genetics course;
integrating a GEP project into the lab of a general genetics course; and
using GEP projects as the core of a “research” or “independent study” course for a small number of students.
Goals, Process, and Outcomes
The primary research goal of the GEP is to improve and annotate the draft quality sequence of large chromosomal regions with the aim of addressing a question using comparative genomics. The current project focuses on the small dot chromosomes from D. virilis, D. grimshawi, D. erecta, and D. mojavensis, as well as large (∼1-Mb) regions from the long autosomal arms of D. erecta and D. mojavensis, selected as euchromatic control regions. The annotation process uses an evidence-based approach for all the finished regions.
The overall process (Figure 2) starts with the publicly available “draft” quality whole genome shotgun assemblies. For the regions of interest, the draft assembly and shotgun reads are obtained from the relevant databases and divided into overlapping student-sized projects. If the assembly is of sufficiently high quality, the finishing step is bypassed (as is the case for D. erecta) and the projects are distributed directly to the GEP member institutions for evidence-based annotation. However, most projects require sequence improvement/finishing before annotation. For finishing, the projects are designed around available fosmids with inserts of ∼40 kb. The fosmids are used as template DNA in sequencing reactions to generate new data. For annotation, projects range from 40 to 60 kb.
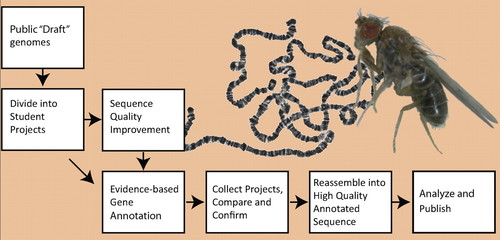
Figure 2. The GEP pipeline. Public whole-genome shotgun assemblies for the genomic regions of interest are split into numerous small “student-sized” projects. In most cases, draft assemblies of the projects are first finished to high-quality (see Materials and Methods for finishing standards). Draft sequences of high quality are given directly to students for annotation. All projects are analyzed by at least two students and then checked for discrepancies between student versions. Finally, the validated projects are reassembled into a single high-quality annotated genomic region for analysis and publication. D. virilis adult fly and a preparation of larval salivary gland polytene chromosomes from D. melanogaster are shown in the background.
Projects are claimed by GEP faculty for distribution to their students. For quality control purposes, two students, usually at different schools, complete each project. After the projects are completed (either finished or annotated), they are submitted back to Washington University through the online Project Management system, which streamlines the comparison of the independent student submissions. When needed, minor adjustments are made to produce a final version; if major disagreements indicate that significant work is needed, the project is returned to the central claim system. Selected alumni of the Bio 4342 course at Washington University work over the summer to reconcile submissions. The validated projects are then assembled into a contiguous genomic region before final analysis of the properties of the region and publication.
Genomic Finishing (Sequence Improvement). Students learn how to use Consed software (Gordon et al., 1998) to manage their finishing project. This software displays the sequence assembly, allowing the student to see any gaps or areas of low-quality data. Students design oligonucleotide primers for DNA sequencing reactions, picking oligonucleotides to address regions of their project with weak or missing data. Students are also asked to specify the type of sequencing chemistry that should be used for a particular reaction. They upload this information to the GEP server. Once a week, all requested oligonucleotides are obtained and used in sequencing by the Washington University Genome Center. The results are then electronically distributed back to the students for incorporation into their projects. Pooling orders and using a 96-well plate format provides considerable cost savings. By using the Genome Center pipeline with this format, a 1-wk turnaround time between oligonucleotide orders and delivery of sequencing results can be obtained. Sequencing is currently available to GEP members over a 10- to 12-wk period each spring semester.
To date, the GEP students have finished and submitted 134 fosmids. This includes fosmids covering the entire banded portions of both the ∼1.2-Mb D. virilis and the ∼1.7-Mb D. mojavensis dot chromosomes. Ongoing finishing work is focused on two large scaffolds that make up a majority of the D. grimshawi dot chromosome and a large ∼1-Mb euchromatic region from one of the long arms of a D. mojavensis chromosome.
Students are made aware of the desired target sequence quality and provided with a “finishing check-list” to enable them to confirm that all standards have been met before submission to Washington University. In addition, students carry out an in silico restriction digest of their finished fosmid for comparison with the digests previously generated from the DNA, as a further check on the assembly. The total amount of improvement can be considerable. For example, in finishing 68 fosmid clones covering 1.7 Mb of DNA from the D. mojavensis dot chromosome, students closed 26 of 28 gaps; added 21,077 base pairs of sequence; and improved many low-quality bases to high-quality standard.
Quality of Student Work: Genomic Finishing (Sequence Improvement). Because each project has been analyzed and submitted by at least two students, we can compare their results as an initial assessment of the quality of the work done. Any regions found to be discrepant between the two sequences were investigated further for errors in finishing. This comparison also allowed us to recognize common errors made by students to further improve the available training materials. A recent analysis showed that of 58 submitted projects, 43 (74%) projects were completely congruent and acceptable, one project had only minor issues with the identification of putative polymorphisms, and 14 projects needed additional data. Most of the errors made by students involved giving undue value to low-quality data.
Genomic Annotation. In annotation, the entire region to be annotated is divided into projects with 40–60 kb of continuous sequence; adjacent projects overlap to maximize the likelihood that each gene in the region is found entirely within at least one project. Students are provided with computer-generated predictions for gene models and other computational analysis as viewed on a local custom copy of the University of California, Santa Cruz, genome browser (Karolchik et al., 2008). Students use these results as well as examination of the well-documented gene models in D. melanogaster to explore putative start and stop sites, as well as all intron and exon boundaries. They base their final gene models on the best evidence available, assuming minimal change from the D. melanogaster gene. The goal is for students to annotate all putative isoforms for each gene found within their particular sequence. Figure 3 shows an example of the results of student annotation. For the gene in question, a putative orthologue of the D. melanogaster gene CG1909 found in D. erecta, the student annotated nine exons, including two short exons at the start of the gene that were not predicted by the four ab initio gene prediction algorithms used. However, the level of conservation of the encoded amino acids suggests that the student-generated model is more likely to be correct than the ab initio predictions.

Figure 3. Example of a student annotation of a gene. (A) Student-generated gene model (orange) compared with models from various ab initio gene prediction algorithms. Note the first two exons (top left) of the manually generated model, not found in any of the ab initio predictions. (B) Alignment of the amino acids of the first two exons of the gene model from D. melanogaster and the student model from D. erecta.
To date, GEP students under the guidance of their faculty advisors have submitted 168 annotation projects back to the GEP. Because each project is annotated at least twice and adjacent projects overlap, most regions are annotated about three times. This represents more than 8 million bases of DNA analyzed. Cumulatively, students have created more than 900 gene annotations. Students rely on the well-annotated D. melanogaster genome (where mRNA/cDNA data are available) to help them predict the isoforms in their projects. This has led to the submission of 1524 gene models. The annotation of the ∼1.2-Mb D. virilis dot chromosome has been completed; annotation of other genomic regions mentioned is currently in progress. The current status of an ∼1.2-Mb region assigned to the dot chromosome from D. erecta is shown in Figure 4. The cumulative coverage shows that 68% of the projects have been analyzed by at least two students, 29% have been covered once, and one project has yet to be analyzed. Other regions currently in progress include a region assigned to arm 3L of D. erecta (60% covered twice, 31% covered once) and the D. mojavensis dot (17% covered twice, 28% covered once).

Figure 4. Current status for the dot chromosome of D. erecta. The top line indicates the status for that section of the chromosome: green, regions that have been submitted back to the GEP at least twice; brown, submitted once; and red, no submissions. Individual clones showing the “golden path” across the region are also color-coded using the same scheme.
Quality of Student Work: Genomic Annotation. Students submit completed data files giving the coding sequence of each of their putative genes, specifying start, splice, and stop coordinates. A detailed “annotation reporting form” is used to report their supporting evidence. Given that comparison with published D. melanogaster gene annotations is an important part of the annotation process, the ability to evaluate student work is complicated by the fact that the annotation of D. melanogaster is being constantly updated. This means that the model best supported by evidence for a given gene in one of the other species may change over time based on new annotation data for D. melanogaster. To assess the quality of the work done by students we concentrated our analysis on 44 genes from the D. erecta dot chromosome whose D. melanogaster annotation did not change over the course of the student work. These 44 genes had 132 different isoforms, and students submitted a total of 338 gene models; of these models, seven (2%) models were judged to have major errors, often because of missing exons. Other errors included incorrect exon length and confusion about the differences between the different isoforms. We observed that students had the most difficulty when they had to reconcile conflicting observations, and they often placed too much value on computationally generated information (e.g., ab initio gene predictions). Overall, 75% of the models submitted were in agreement with others for that gene and passed our quality control checks.
Student Outcomes
Knowledge-based Outcomes.Figure 5 represents the outcomes for the pre- and postcourse knowledge-based quizzes given during the 2008–2009 academic year. For annotation, the results show a significant improvement (p < 0.05) of the mean score for students who participated in annotation projects and for students who performed both annotation and finishing projects. These scores were significantly higher (p < 0.05) than those of the comparison group of students who did not use GEP material in their studies (Figure 5). The comparison group test means did not improve from pretest to posttest. For finishing, the group that participated in finishing and annotation projects had a higher mean score (p < 0.05) than all other groups. Groups that did not do finishing projects did not differ from each other or from the precourse results.
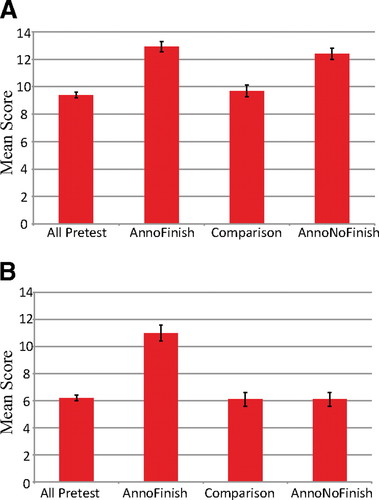
Figure 5. Student quiz score results. The mean quiz score results show a significant increase in average test scores for GEP students who participated in the relevant project. The left column shows the average precourse score for all groups. The AnnoFinish group is made up of those students who participated in both an annotation-based project and a finishing-based project. The comparison group was the group of non-GEP students recruited at GEP-member schools (see text). The AnnoNoFinish group is made up of those students who participated in an annotation-based project but not a finishing-based project. (A) Annotation quiz (genes and genomes). (B) Finishing quiz (sequencing reactions and data analysis). Error bars represent 2 SEs above and below the mean.
Faculty who use the GEP projects report additional student impacts that are not captured in a knowledge-based survey. We collected these observations and found several common themes (Table 2). An important aspect of the GEP approach is to give students an introduction to the process of research and an understanding of how new knowledge is created in the field. As reported previously (Lopatto et al., 2008), students in GEP-associated courses gained many of the skills and attitudes normally associated with spending a summer doing research. Faculty report that students gained skills in critical analysis and a realization that computer-based evidence can be incorrect. Students also learned to cope with obstacles faced in the research process. Furthermore, faculty report that active problem solving solidifies understanding, grappling with the unknown builds confidence, and contributing to a larger project builds pride and a sense of significance of effort. The observations of individual faculty are reported in Table S1 (see Supplemental Material), color-coded to the themes listed in Table 2.
As part of the attitudinal survey, we asked students to assess how much they gained from the various teaching tools and course activities. These included the various training materials as well as activities more closely associated with research, such as preparing oral and written reports on their research and defending their conclusions. Figure 6 summarizes student responses to 18 different components. Students consistently reported more gains from their own research project than from problem exercises designed to teach the use of the computer tools. For example, the highest reported gains were assigned to the actual finishing and annotation activities, whereas training material and exercises were given lower scores.
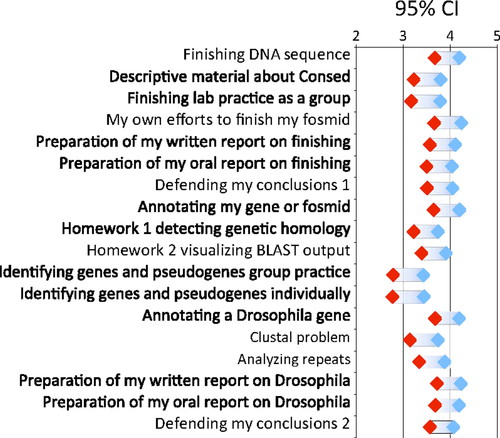
Figure 6. Student ratings of various GEP activities. Plotted are one sample confidence intervals (n = 77) for 18 items. Students rated each activity to indicate how much it contributed to their learning experience based on a 1 (little learning) to 5 (very beneficial) scale. The “Defending my conclusions” was asked once at the end of the annotation questions (1) and again at the end of the finishing questions (2).
Interestingly, the results of the student-perceived gains derived from the various activities show a positive correlation with the postcourse quiz scores. Those students who reported the most benefit from these activities showed a higher average score on the annotation questions of the quiz. Figure 7 shows the relationship between the benefit the student assigned to the activity of “annotating my gene/fosmid” and the average quiz scores. Analysis of the data here indicates an overall significant difference between groups (p < 0.05). Many of the activities listed in Figure 6 showed similar correlations.
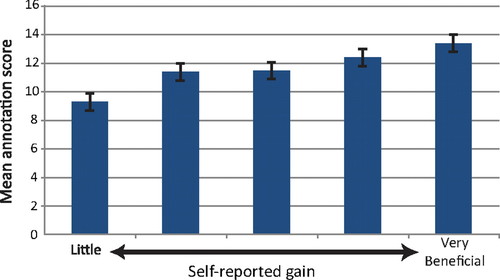
Figure 7. Mean quiz score separated by self-reported gains. Groups of students were separated based on how much benefit they reported from “Annotating my gene/fosmid” on a 1 (little learning) to 5 (very beneficial) scale. The mean annotation quiz score is reported for each group. The overall analysis of these groups reveals significant differences (p < 0.05). The error bars represent 2 SEs above and below the mean.
Attitudinal Outcomes. Students were asked to respond to a series of questions that asked them to rank their response to various aspects of their GEP experience on a 1 (strongly disagree) to 5 (strongly agree) scale. Data from this survey were collected in both the 2007–2008 and 2008–2009 academic years. In the precourse survey, these questions focused on such aspects as reasons the student chose the course, their self-perceived ability to undertake various course activities, and their attitudes about themselves and science. In addition to the above-mentioned points, postcourse surveys asked the students to rate how much they gained as a result of taking the class, reflecting on their own learning behaviors.
Figure 8 shows the cumulative results for GEP 2008 and GEP 2009 students in three different areas and compares them to results from a previous survey of students who worked for the summer in a research lab and took a similar survey (the SURE follow-up survey; see Lopatto, 2008). In all cases, two-thirds of the students either “agreed” or “strongly agreed” with the statements that taking the course with GEP research “helped them to become a more active learner,” “helped them learn to think independently,” and “increased their motivation to learn.” The results of these attitudinal surveys compare very well with the results for students who responded to similar questions after they participated in a summer research project.
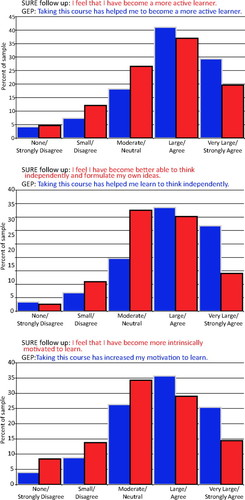
Figure 8. Student-reported gains comparing a summer research experience (SURE) with a GEP academic year course (GEP). These graphs show the cumulative percentages of GEP and SURE respondents from both the 2007–2008 and 2008–2009 academic years. The students in each survey were responding to slightly different statements, shown in each chart. (Top) Thinking independently. (Middle) Motivation to learn. (Bottom) Active learner. The SURE statement is shown in red and the GEP statement is shown in blue.
We also asked students whether their plans for future education were impacted by their participation in courses with GEP activities (Figure 9). Although most of the students had plans for postbaccalaureate education that did not change as a result of their GEP experience with research, for others the experience did have an influence on their long-term plans. Some students (8.3%) reported that the experience had confirmed for them plans to pursue postbaccalaureate education. Another 5.0% of students reported that although they did not have a plan before taking the class they now did; this group was in contrast to an equal-sized group that decided to drop such a plan at the end of the semester in which they took a GEP-affiliated course. Several students (9.4%) reported that after taking the class their plans for postbaccalaureate education now included a greater emphasis on genomics.
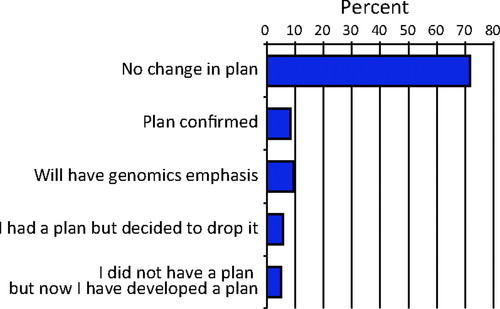
Figure 9. Impact on student plans. Each student was asked about his/her plans to pursue postbaccalaureate education. Percentages of respondents for each category are shown. These are the averages for the 2007–2008 and 2008–2009 GEP student surveys.
As part of the online survey, we allowed space for open-ended comments. Almost half (48.8%) of all students provided comments in the 2009 survey. Many of these comments indicated that students were experiencing both the positive aspects of research as well as the frustration that comes with working with the unknown. Here are some examples that highlight this theme:
“… the use of novel research for the laboratory … brought meaning and insight into what we were learning in class.”
“The class was very intellectually challenging for me. It taught me to think in a way that I had never thought before.”
“This type of work forced me to think critically and creatively about DNA …”
“It's very frustrating when you don't have an expert right next to you to answer questions.”
“I learned to fight through the frustration and eventually figure out the problem.”
“The homework and packets should be more explained, there was too much ambiguity with some of the steps.”
“Several times throughout the semester I felt like we were just given a task with no real instruction on how to accomplish it. This could be due to a miscommunication between the professor and I, but I still feel as if I spent a lot of time playing catch up …”
“I guess we learned about genomics through doing the tasks that we were assigned; it was sort of a self-teaching class. While I found I certainly learned a fair amount about genomics, I would have liked a bit more guidance with a few of the steps.”
“Our professor didn't give us any tools to learn. Everything we gained from the class to help us annotate the sequence was either found by desperately messing around on the various websites thrown at us or by talking with other students.”
Faculty Outcomes
Although the primary focus of the GEP is to bring genomics education to undergraduates, one of the extra benefits is enhanced faculty development and networking. Many of the GEP partners belong to small schools with no other faculty working in genomics and insufficient financial resources to undertake student-led sequencing projects. GEP members support each other by maintaining a culture of training, assessment, and research, as well as being a support network for both new and tenured faculty. To assess the impact membership in the group has provided, we asked all faculty members to answer an online survey of professional gains. Responses were collected by Washington University computer staff and de-identified before forwarding to D.L. for analysis. Forty-eight faculty (of 50 eligible) voluntarily took the survey and the cumulative results are shown in Figure 10. The respondents were asked on a 1 (strongly disagree) to 5 (agree) scale if they felt that the GEP helped them on 15 different items. On all but one item the average faculty response was very positive, with a mean between 4 and 5. The one question receiving a lower response was #13, “Other faculty in my department express interest in applying this approach in their own area.” The strongest levels of agreement were for questions 7) “Overall, I had a positive experience with GEP” and 8) “I plan to use the GEP program again.”
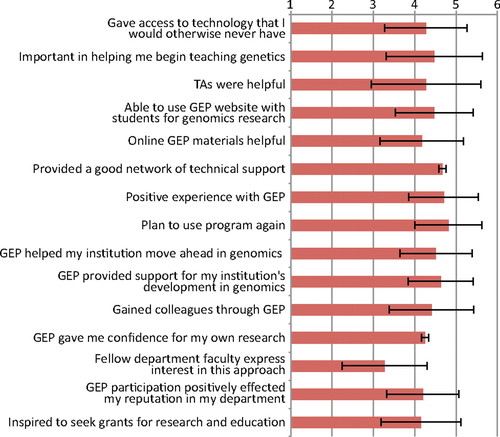
Figure 10. Faculty survey results on the impact of GEP participation. Faculty responded on a 1 (strongly disagree) to 5 (agree) scale to various statements. Mean and 1 SD are shown.
DISCUSSION
Keys to Success
During the development and growth of the GEP program, we have identified several themes that empower a continued, successful partnership, because collectively we have learned important lessons regarding the implementation of a GEP-style research project within an academic year class. An important principle in the GEP system is the idea of “common tools, different problems.” The techniques and analytical approaches students need to learn to improve draft sequence and/or annotate genomic sequence are basically the same for all, but each student has a unique project. This allows the instructor to teach the students as a group, while each student gains a sense of “ownership” for their project, even though all students are doing basically the same sort of investigation. The “common tools” also allow for peer teaching—students are often able to help each other with technical issues that crop up when performing the analysis. Different challenges arise in different projects, but there are also common stumbling blocks; once one member of the class has dealt with such an issue, he or she can share the newly developed expertise. Students also appreciate contributing to a larger project.
The availability of centralized technical support plays a crucial role in introducing GEP-style activities on new campuses and forms the fulcrum on which the distributed model is able to function. Figure 10 indicates the strong consensus faculty have about the importance of centralized contiguous technical support. The web-based system to manage curriculum materials, provide student-ready projects, and provide timely technical support helps faculty use class time effectively.
We also found it important for a successful class experience to have sufficient lab time at any one sitting for students to do computer-based analysis. Students found that it took some time to “get up to speed” with an analysis, and anything less than a block of 2 h gave students too little time to make good progress on their project. Computer-based analysis does not pose the same safety issues as wet lab-work, so more flexibility is possible as to when and where students work. However, although students have worked on a GEP project in isolation, in practice working in groups during assigned class periods provides many benefits. Analyzing problems as a group helps keep up momentum, stimulates peer instruction, and leads to a group dynamic that helps overcome the sometimes tedious task of annotation. Peer instruction is frequent in GEP classes, because these classes often attract a mixture of computer-savvy and not-so-savvy students. Nonetheless, we find it advantageous to have one experienced person (faculty or TA) per six to seven novices. Given the open-ended nature of annotation projects, regular meetings with set mileposts are also important to keep track of students' progress. This allows students to pace their work over the allotted portion of the course. Insight by many of the member faculty on implementation in different settings and the lessons learned are provided in Table S2 (see Supplemental Material).
An added value of the GEP has been the collegial support of a group of college and university faculty with shared goals. The group effort is sustained by summer “Alumni Workshops.” These workshops are short events usually scheduled over a weekend (Friday evening to Sunday afternoon). Such meetings allow for updated technical training to keep the faculty current on new computational tools within the GEP computer infrastructure. They also help create and strengthen connections between members, allowing members to share curriculum and ideas, as well as design joint assessment tools, plan future scientific and pedagogical efforts, and work on both scientific and pedagogical manuscripts for publication. Interinstitutional interactions of this type have been recognized as an important mechanism for promoting faculty development and student learning in research (BIO 2010).
Challenges on Implementation
Implementation of the GEP curricula requires dealing with several challenges, which vary with institutional context. In spite of the different specifics, a set of common issues emerges. One of the challenges of bringing advanced genomic training to undergraduates is overcoming student attitudes toward learning: many students are geared toward lecture classes, memorization of facts, and “canned” laboratory courses with known outcomes. Approaching coursework that involves novel research is a new experience for most students. Some initially consider the challenge “unfair,” given their status as undergraduates, but most have eventually been convinced by their own success in making novel contributions. Another challenge is that some faculty and students do not see computer-based analysis as “real research.” We see annotation as a series of in silico experiments—each algorithm used to analyze a sequence, whether an ab initio gene prediction or a BLAST search for conservation, produces evidence as to the likely gene model. Students are asked to integrate and analyze all of the evidence produced by these tools, determine the best gene model based on this evidence, and be prepared to defend their decisions. We view the gene model as a hypothesis supported by evidence.
Other difficulties include the ever-present technical and software issues. Consed is limited to the Linux or Mac OSX platforms, which are often not supported by the technical staff at member schools. Similarly, the operating system used varied greatly among the partners. Although the GEP-developed methods have been designed to work across many different computer platforms, issues continually arise as new members join the project and try to make the projects work with their pre-existing computer facilities. Thus access to knowledgeable information technology support, provided either centrally or locally, is essential. Another challenge in the GEP curriculum is coordinating sequencing reactions. To keep class work on schedule (within the constraints of a quarter or semester course), we aim to achieve a 7-d turnaround between the student request, specifying the oligonucleotides to be ordered, and the posting of sequencing results. This allows little margin for error or delay. Requests are accepted once per week (Wednesday midnight), which is not ideal for a Tuesday/Thursday class. Faculty proved adept at adapting their course schedules accordingly. Finally, the unpredictable nature of experimental work must be an expected feature of the course. Not all problems are solved on schedule, so grading must to some degree reflect the quality of the student's analysis rather than demanding complete resolution. As a consequence, completion of large assemblies (e.g., the 1- to 2-Mb dot chromosome of a given species) generally stretches over 2–3 yr.
Future Goals
It is clear that as DNA sequencing gets cheaper, more data will be available for analysis. The wealth of publicly available genomic data makes projects like those undertaken by the GEP possible by removing the financial barriers created by the high start-up costs of high-throughput sequencing. The ability to sequence is far out-stripping the ability to analyze, leaving plenty of room for student projects, and an increasing need for student–scientist partnerships. Furthermore, sequencing costs may soon be low enough that a consortium such as the GEP could accomplish a project requiring de novo sequencing for a reasonable cost. By using a common set of techniques, it is possible for students to make significant contributions to the analysis of large data sets by a “divide and conquer” strategy. A long-term goal of the GEP is to provide the necessary training and infrastructure to allow faculty a wider variety of interesting species and/or gene families to be analyzed. We also anticipate opportunities for international collaborators and community colleges to participate.
Genomics is particularly well-suited for a distributed research project based on student–scientist partnerships: 1) projects can be readily broken down into comparable-sized chunks; 2) the necessary hardware is readily available; 3) the necessary analytical tools are accessible to undergraduate students while still challenging them, making the effort pedagogically valuable; and 4) the individual projects can be reassembled to make a more meaningful whole. Several other national projects that use genomics or other high-throughput analysis in undergraduate research have also been reported. Several members of the GEP faculty have also been active members of the Genome Consortium for Active Teaching (GCAT; www.bio.davidson.edu/projects/GCAT/GCAT.html) led by Malcolm Campbell of Davidson College (Campbell et al., 2006). GCAT allows students to participate in research using DNA microarrays. GCAT has provided training workshops for faculty covering both wet bench techniques and computational analysis for DNA microarray experiments. GCAT also provides arrays and access to an array reader to collect student-generated data. In this program, faculty defines the research goals for the individual projects. This differs from the GEP, where all members are working on a single larger research question. Similar to the GEP results reported here, the GCAT program is an effective way of engaging students in genomics and developing an interest in research (Campbell et al., 2007).
The GEP partnership in many ways follows the successful pattern established by GCAT, albeit using different technologies. As a group, GEP members recognize the following principles:
The clear need to bring genomics into the undergraduate curriculum
A focus on research problems that can lead to scientific publication
Development and sharing of teaching materials, thereby maintaining a flexible approach, so that all schools can participate
Development of a distributed community of faculty/student researchers
Making science education research (assessment) part of our joint effort
Fewer projects focus on eukaryotic genomes, which present additional problems of data management, as well as being more challenging to annotate. The Dolan DNA Learning Center, Cold Spring Harbor Laboratory, runs the Dynamic Gene website at www.dynamicgene.org. This site provides students with tutorials on gene annotation and has a web-based annotation system that students can use to annotate a region of the rice genome (Miklos et al., 2006; Hacisalihoglu et al., 2008). However, unlike the GEP system that is collecting the student work for eventual analysis and publication, the Dynamic Gene website does not as yet seem to collate, check, and use the student work done on the rice genome. Partnerships similar to the GEP could be created for a variety of model organisms. Projects amenable to this type of distributed analysis could include any species for which draft sequence and mapped fosmids are available, where the scientific problem is of sufficient interest, and the research community is tolerant of the slower pace of analysis.
In summary, the GEP has provided hundreds of students across the country a means to engage in meaningful twenty-first century research. The GEP has proved itself a flexible partnership, made up of a wide variety of institutions, whose members have succeeded in bringing genomics research experiences into the undergraduate curriculum. This has resulted in both documented benefits to our students and in bona fide scientific discoveries. Although advances in sequencing technology continually force us to reconsider how best to incorporate these experiences into our teaching, we believe interaction among the GEP faculty and the faculty and staff at Washington University will continue to be a fruitful source of ideas into the future. Clearly, the ongoing need for annotation of new sequence data and the benefits of partnerships such as the GEP for students and the larger scientific community bode well for the continuation of such a strategy.
ACKNOWLEDGMENTS
We appreciate the materials received through the Drosophila Genomics Resource Center. We thank the staff of the Washington University Genome Center for continuing assistance. Special thanks to Frances Thuet for managing the assessment web pages and to Jeannette Wong for collecting data on common errors while reconciling student projects. This work was funded in part by HHMI grant 52005780 and National Institutes of Health grant R0I GM068388.



