WWW.Cell Biology Education: Using the World Wide Web To Develop a New Teaching Topic
Cell Biology Education calls attention each quarter to several Web sites of educational interest to the biology community. The journal does not endorse or guarantee the accuracy of the information at any of the listed sites. If you want to comment on the selections or suggest future inclusions, please send a message to E-mail: [email protected]. The sites listed below were last accessed on February 21, 2005.
The Internet provides access to an enormous array of potential teaching materials. Below, we describe one approach for using the World Wide Web to develop a new college biology laboratory exercise. As a topic example for the search strategy, we have selected signal transduction.
INTRODUCTION
Evolt http://browsers.evolt.org/
There are two essential ingredients for collecting information from the Internet: 1) a World Wide Web (Web) browser, and 2) an Internet search engine. The two most widely used Web browsers are Microsoft's Internet Explorer and Netscape Communications Corporation's Navigator. There are, however, more than 120 browser software possibilities available. These browsers have been cataloged by Adrian Roselli and may be found at Evolt, a site that supports the exchange of Web design information. Web browsers are like brands of shoes; some fit better than others. By visiting Evolt, whose uniform resource locator (URL) is listed below, one might find a browser that works better. For example, some Internet users prefer Firefox (Figure 1) and Safari, because Web browsers have different efficiencies in displaying Internet information on different computer platforms.
As is the case with Web browsers, there are a number of Internet search engines. Each search engine has a variety of features that address different objectives for finding information from the Internet. The State University of New York at Albany has organized an excellent list of Internet search engines.
University Libraries Internet Search Engines http://library.albany.edu/internet/engines.html
The Albany site also includes a guide on “How to Choose a Search Engine or Directory.” More than 80 search engines are placed into categories ranging from Meta Search, Deep Web, and Domain Names, to FTP Search. The choice of Internet search engine resembles the decision of which library one wishes to visit. Some libraries may have more books, others have better photo archives, and some are simply more comfortable to sit in. Currently, the most widely used Internet search engine is Google. Other popular search engines include HotBot, Lycos, AltaVista, and Dogpile.
For the purpose of this report, we will use Internet Explorer as the Web browser and Google as the initial Internet search engine. Most college undergraduates are thoroughly familiar with both.
THE INITIAL SEARCH
We begin our initial research into Internet resources for developing a new lab on signal transduction by using the Internet search engine Google.
Google http://www.google.com/
Google is a California-based company whose corporate name refers to the number known as a googol: a 1 followed by 100 zeros. The organization was started in 1998, and its prominence today as an Internet search engine is clear.
The term “signal transduction” is typed into the search box with quotes to limit the search to this exact two-word phrase (Figure 2). This query results in 1,760,000 returns (or “hits”), which is clearly far too many possibilities to be worthwhile. Google has a feature that will allow the search to be narrowed with a category called “Search within the results.” Into that box we typed “lab exercise” in quotes. These two combined search phrases result in 94 hits. The first listing of the 94 returns is to an article titled “Signal Transduction and Control of the Cell in Yeast, Saccharomyces cerevisiae: A Collaborative Laboratory Exercise.” Google provides a path to that article.
Figure 1. Search page from the Web browser known as Firefox. Image used with permission of Mozilla, parent of Firefox. For a free copy of Firefox, which operates on either Apple or IBM compatibles, visit http://www.mozilla.org.
Association for Biology Laboratory Education Archives http://www.zoo.utoronto.ca/able/volumes/vol-19/05-hoopes/05-hoopes.htm
This complete lab exercise was presented to the 1998 Association for Biology Laboratory Education (ABLE) meeting by Barbara Hoopes, Nancy L. Pruitt, Kathleen Baier, and Sherry Brooks of Colgate University. The Google-located reference is on target as a lab exercise based on signal transduction. ABLE is an organization that meets annually for the purpose of sharing tested lab exercises among the participants. The organization's Web site is found below.
ABLE http://www.zoo.utoronto.ca/able
Dr. Hoopes indicated that the figure used in the exercise is out of date (see Figure 3). She also indicated that the lab exercise was in need of revision because of advances in the field. She also had a very interesting observation. It is harder to get innovative lab exercises into print today, she argues, because such exercises must be accompanied by extensive assessment documentation. For lab developers at smaller institutions, such documentation is prohibitively expensive (B. Hoopes, personal communication).
An examination of the other 93 Google returns results in very little, for the phrase “signal transduction” is frequently used in course syllabi and in vitae. Thus, from the 94 hits, one viable resource was located for a possible lab exercise.
Figure 2. Google is the trademark of Google, Inc. The image shown is the famous DNA logo celebrating the 50th anniversary of the 1953 article on the structure of DNA. The phrase “signal transduction” is entered into the search box.
Figure 3. Title page of the lab exercise as available at the ABLE Web site along with the first figure from the article illustrating a principle being explored by the lab exercise. The image is used with the permission of Dr. Hoopes, whose important comment is found in the text.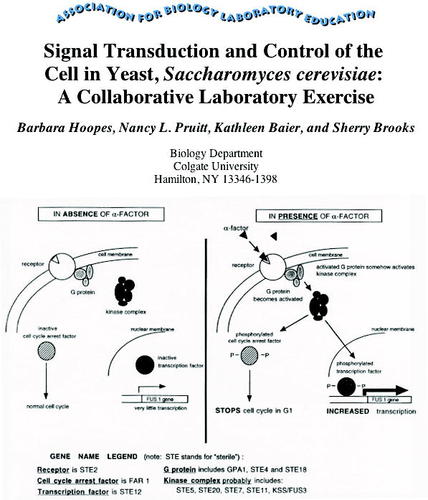
THE SECOND ITERATION
This Google search can be considered to be both successful and unsuccessful: one good reference and only one reference from the whole of the Internet. A different search strategy might identify more Internet resources. Perhaps different key terms for the search might yield better results. What terms should be used instead of “signal transduction”?
We suggest turning to a respected Internet biology textbook, Kimball's Biology. John W. Kimball has converted his 1994 general biology textbook into digital form. The original text is continually updated and expanded.
Kimball's Biology Pages http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/
Typing the term “signal transduction” yields no returns; however, “second messengers” and “cell signaling” do. Topics under cell signaling include NO receptors, steroid receptors, G-protein-coupled receptors, cytokine receptors, and TGF receptors. Second messenger topics include cyclic nucleotides, inositol trisphosphate, and calcium ions. Although no lab material is found at Kimball's textbook site, both topics found here serve as an excellent reference for students being introduced to signal transduction. Kimball's topic selection also suggests that a return to Google might yield more by using “second messenger” and “cell signaling” as search terms.
Another source of general information may be found at the URL listed below.
Wikipedia, The Free Encyclopedia http://en.wikipedia.org/wiki/Main_Page
This Internet-based encyclopedia was initiated in 2001 by Jimmy Wales and Larry Sanger. Today the site has over 1,300,000 articles from more than 13,000 contributors. This reference has information on just about any topic imaginable. The search term “signal transduction” yields a detailed article on the topic that has key terms “hot-linked” to other articles in the encyclopedia (Figure 4). Information is provided about extracellular, intracellular, and intercellular signaling. Transmembrane, nuclear, steroid, and even orphan receptors are described. Signal amplification is briefly mentioned. The entry concludes with a short bibliography and external links. The site, however, must be used with caution, for the articles are “user” edited. Undergraduate students would find this a good site for background information on signal transduction, but because of the lack of professional editing, the site should be cited in articles with caution.
Figure 4. Wikipedia is a free, user-developed encyclopedia that is also user edited. Permission has been given to use the image with proper notification of source.
A metasite (a site that lists links to other sites) can be found at The WWW Virtual Library, the oldest metasite on the Internet, founded in 1991. One of its divisions is Biochemistry and Cell Biology, which is maintained by Gabriel Fenteany of the University of Illinois at Chicago.
The Virtual Library of Biochemistry and Cell Biology http://www.biochemweb.org/signaling.shtml
This Virtual Library of Biochemistry and Cell Biology (VLBCB) has a section specifically oriented toward signal transduction information. The links found here include The Medical Biochemistry Page, Dynamic Signaling Maps, The Signaling Pathway Database, and 10 receptors and signaling effector sites. One exceptional resource listed here is to the Sigma-Aldrich Chemical Company, where one can download over 80 free, remarkable graphics representing cell signal pathways that may be used for instruction (Figure 5). The main VLBCB page concludes with a listing of nearly 40 research labs that are focused on signal transduction research. This site would serve to educate the college instructor as well as the student; however, it does not list any lab exercises on signal transduction.
Sigma-Aldrich Cell Signaling Pathway Slides and Charts http://www.sigmaaldrich.com/Area_of_Interest/Life_Science/Cell_Signaling/Scientific_Resources/Pathway_Slides___Charts.html
Figure 5. One of more than 80 digital posters available at the Sigma-Aldrich site. The posters, in PowerPoint format, may be downloaded free for personal use or for educational presentations. Sigma-Aldrich is a producer and distributor of chemicals with corporate offices in St. Louis, MO.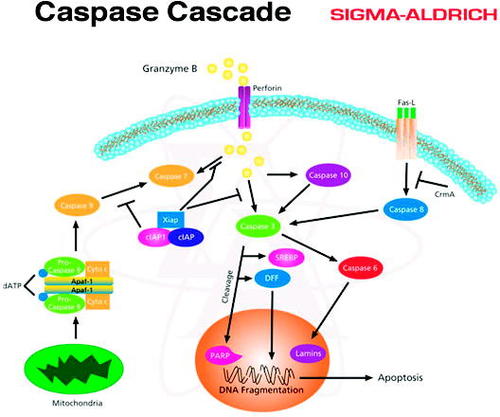
A FINAL ITERATION
Several groups with significant financial support have developed Web locations of science teaching information. One of the more established sites representing this effort is MERLOT.
Figure 6. A screen shot of the MERLOT return information box for the search on the phrase “signal transduction.” Image used with permission.
MERLOT http://www.merlot.org
MERLOT, a term representing Multimedia Educational Resource for Learning and Online Teaching, was founded in 1997 by the California State University Center for Distributed Learning. It is an open-source collection of nearly 4,000 Web-based learning sites and materials. In 1998, MERLOT expanded to include the University of Georgia System, Oklahoma State Regents for Higher Education, and the University of North Carolina System. By 2000, 23 educational systems were incorporated into MERLOT. Individuals can also join MERLOT at no expense, and there is an extensive membership directory with over 3,100 members in the science and technology area.
By typing the phrase “signal transduction” into the prominent search box at the top of the MERLOT homepage, three returns are obtained. The first is authored by Joyce J. Diwan of Rensselaer Polytechnic Institute (Figure 6). The teaching resource is extensive, and the section on signal transduction has excellent learning materials, including three animations. The second listing is for teaching materials developed by Larry L. Keeley of Texas A&M University. One of his animations focuses on G-protein signaling. The third represents a collection of materials by Yu-Wai Peter Lin of Barry University. It consists of information dealing with Nobel Prizes in physiology and medicine dealing with membrane-based signaling events. Using the phrase “cell signal(l)ing” results in two more hits, both of which contain animations.
The MERLOT site is well conceived and executed. Each reference clearly identifies the URL, the author, and the source. Each resource outlines requirements for use. Most resources have been rated as to content quality and potential effectiveness as a teaching tool. There is also an ease-of-use assessment. However, a laboratory exercise on signal transduction is not among its holdings.
Figure 7. The logo for the NSDL. Permission to use the logo is granted on the Web site.
Our last stop is at the National Science Foundation-sponsored National Science Digital Library, also known as NSDL. The Mission Statement of the NSDL states: “NSDL provides educational resources for science, technology, engineering, and mathematics education. The NSDL mission is to both deepen and extend science literacy through access to materials and methods that reveal the nature of the physical universe and the intellectual means by which we discover and understand it.” This educational resource opened in December of 2002, and it continues to grow (Figure 7).
National Science Digital Library http://nsdl.org/
NSDL's simple homepage has a search box near the top, not unlike MERLOT. When “signal transduction” is entered, 245 records are retrieved. Unlike Google, these hits are all very pertinent, including Science's Signal Transduction Knowledge Environment (STKE). STKE is a weekly electronic publication of current events in signal transduction research and is a joint venture between the American Association for the Advancement of Science (AAAS) and Stanford University. A portion of STKE is public, and the rest can be accessed through AAAS membership. Regular visits to this site would keep an instructor up to date as to developments in cell signaling.
STKE: Signal Transduction Knowledge Environment http://stke.sciencemag.org/
Currently, the NSDL has over 420 collections of educational information ranging from “All about Birds” to “The Learning Matrix.” Like a good book, it is difficult not to browse through the Science Library. When the term “lab” is added to the search strategy, six hits result, with two being labs demonstrating signal transduction. The Hoopes et al. yeast lab mentioned earlier is one of the two. As resources are identified and cataloged, they are being added to this database.
IN CONCLUSION
Signal transduction is a major research topic today. Surprisingly few teaching lab exercises exist that provide undergraduate students hands-on exposure to the topic. We knew that teaching material in this area would be difficult to find, and thus the topic served us well in defining a strategy for locating teaching material using Internet resources. As one identifies the need to locate new teaching material, changing “signal transduction” to some other key term should lead to useful references using the strategy outlined here.



