A Kinesthetic Model Demonstrating Molecular Interactions Involved in Anterior-Posterior Pattern Formation in Drosophila
Abstract
Prerequisites for the Developmental Biology course at Augustana College are introductory courses in zoology and cell biology. After introductory courses students appreciate the fact that proteins have three-dimensional structures; however, they often fail to recognize how protein interactions with other cellular components can lead to specific cellular responses. One of the first topics covered in Augustana's Developmental Biology course is anterior-posterior axis determination in Drosophila. In the past, the subject was taught with a series of graphs demonstrating mRNA and protein concentrations along the anterior-posterior axis. However, this pedagogy was too conceptual for the majority of students enrolled in the course. To aid their understanding, a kinesthetic model of the molecular interactions involving bicoid, nanos, hunchback, and caudal transcripts and proteins utilizing colored pipe cleaners and beads was created. Students model molecular interactions between proteins (beads) and transcripts (pipe cleaners) by placing the appropriate bead on the appropriate pipe cleaner. After working with the model, the concept of molecular interactions became more concrete to students, and they were able to conceptualize anterior-posterior axis determination in Drosophila more clearly. Throughout the rest of the course, students were able to understand molecular interactions without the aid of additional models.
INTRODUCTION
Developmental Biology at Augustana College is a 300-level course requiring two introductory courses as prerequisites, Introductory Zoology and Introductory Cell Biology. After completing the Cell Biology course, students have a general understanding of cellular organelles, metabolism, DNA replication, transcription, translation, protein structure, membranes, and the cell cycle with minimal exposure to cell signaling, apoptosis, and protein trafficking. Aside from a basic understanding of enzyme/substrate interactions, students fail to appreciate the importance of molecular interactions that occur within cells. This creates a challenge when understanding molecular interactions is integral to understanding developmental biology.
Augustana's Developmental Biology course begins with discussions of early developmental events (cleavage, axis formation, and gastrulation) in model systems including Drosophila, Xenopus, chick, and mouse followed by the topic of organogenesis within mammalian model systems. One of the first topics covered in the course is anterior-posterior (A/P) axis development in Drosophila. After completing the course, students often commented that the most challenging part of the course was Drosophila A/P patterning. On exams, they had difficulty explaining in their own words how the A/P axis is patterned during early Drosophila development. Essay questions included on exams indicated approximately one-third of the students in the course did not understand the difference between transcriptional and translational regulation of gene expression. Many students did not appreciate the difference between maternal and zygotic transcripts, and many failed to see the interrelatedness of multiple protein gradients established in the early embryo. Furthermore, approximately 50% of students had difficulty identifying the underlying cause of mutant phenotypes presented on exams. Performance on exams indicated some students understood A/P patterning only at a superficial level, and they could not apply their knowledge to novel problems. This evidence suggested alternate pedagogies might enhance student learning of this complex topic.
BRIEF OVERVIEW OF A/P PATTERNING OF EARLY DROSOPHILA EMBRYOS
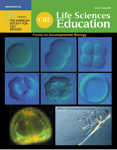
Developmental biology textbooks often describe early A/P patterning in Drosophila in terms of four important protein gradients (Wilt and Hake, 2004, figure 3.18, p. 54; Gilbert, 2006; Wolpert, 2007, figures 2.11 and 2.12, pp. 43–44). After initial patterning is established, GAP, pair-rule, segment polarity, and homeotic selector genes further refine the pattern. The four early gradients include Bicoid, Hunchback, Caudal, and Nanos proteins. Bicoid and Hunchback are required for the development of anterior structures, whereas Caudal and Nanos are required for posterior development. In early A/P patterning of Drosophila embryos, translational regulation is just as important as transcriptional regulation. Maternal transcripts, many of which are localized to specific regions of the ooctye, are deposited in the oocyte during oogenesis. bicoid transcripts are localized in the anterior end of the ooctye, nanos transcripts are localized in the posterior end of the oocyte, and hunchback and caudal transcripts are evenly distributed throughout the cytoplasm of the oocyte. On fertilization, bicoid transcripts are translated to produce an anterior-to-posterior concentration gradient of bicoid protein. Bicoid interacts with caudal transcripts to block translation of caudal transcripts in the anterior end of the embryo, where the two molecules colocalize. Thus, the Bicoid gradient has established an opposing Caudal gradient whose highest concentration is found in the posterior end of the embryo. At the same time, nanos transcripts are being translated to produce Nanos protein. Nanos indirectly interacts with hunchback transcripts repressing their translation. Simultaneously, Bicoid acts as a transcriptional activator regulating expression of the zygotic hunchback gene. As a result of Bicoid activity, zygotic hunchback transcripts are localized in the anterior end of the embryo. The activities of Nanos and Bicoid result in an anterior to posterior gradient of hunchback protein in the embryo. To the trained eye, these molecular interactions that establish opposing protein gradients can be visualized in graphs (Figure 1). Students often memorize gene names and curves on a graph but fail to appreciate the complexity of the process.
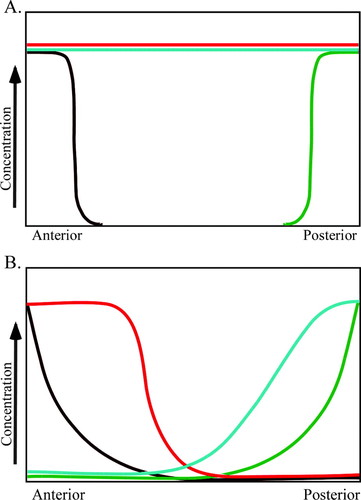
Figure 1. Concentration gradients of maternal mRNAs and zygotic proteins. (A) Concentration gradients of maternal mRNAs deposited in the developing oocyte. (B) Concentration of zygotic proteins as a result of molecular interactions in the syncytial blastoderm. Colored lines in A and B represent transcripts and proteins, respectively: bicoid (brown), nanos (green), hunchback (red), and caudal (aqua). Both A and B are fashioned after Gilbert (2006), figure 9.12, p. 272.
To help students visualize and understand molecular interactions without the use of technically demanding protocols, expensive equipment, or complex computer programs, a “low-tech” kinesthetic activity that models the interactions between four molecules in early A/P patterning in Drosophila was developed. The goal of this activity is for students to use a constructivist approach to understand how molecular interactions in the newly fertilized Drosophila ooctye establish gradients of proteins that pattern the A/P axis of the embryo.
OBJECTIVES OF PIPE CLEANER/BEAD ACTIVITY
The objectives of pipe cleaner/bead activity are as follows:
Improve student understanding of molecular interactions in cells
Improve student understanding of how one molecular gradient can influence others
Improve student understanding of how mutations affect A/P patterning in Drosophila
THE ACTIVITY
Before the class meeting on the day of the activity, students have read textbook sections covering A/P patterning in Drosophila embryos. The reading assignment provides a familiarity with gene names and a basic idea of the hierarchy of genes involved in the process. Before starting the activity, a short lecture covering the morphology of a developing Drosophila embryo is presented.
Handouts detailing the modeling activity's instructions are distributed to students with a packet containing pipe cleaners and beads (Table 1 and Figure 2). Students' first experiencewith the activity is instructor guided and takes approximately 45 min. Periodically, as students are guided through the exercise, molecular interactions are reviewed by drawing graphs that represent the concentration gradients of the transcripts and proteins that have been discussed. At the end of class, students have the opportunity to “check-out” their packets to practice at home. The model is used a second time during a 2-h discussion to demonstrate the effects of mutations on A/P patterning.
| 8 | 2-inch-long brown pipe cleaners |
| 8 | 2-inch-long aqua pipe cleaners |
| 8 | 2-inch-long green pipe cleaners |
| 8 | 2-inch-long red pipe cleaners |
| 4 | 2-inch-long striped red pipe cleaners |
| 8 | Brown beads |
| 8 | Aqua beads |
| 8 | Green beads |
| 8 | Red beads |

Figure 2. Activity instructions distributed to students. Students receive a detailed instruction sheet as well as a blank sheet of paper to complete the activity.
In reality, A/P patterning is a very complex process involving numerous genes (Nüsslein-Volhard, 1991; Brody, 2007; FlyMove [http://flymove.uni-muenster.de/Processes/AxisSpec/AxisSpecEarlyEmbryo/Anteriorposterior/Anteriorposteriorpage.html] as described by Weigmann et al., 2003). In this activity, students focus on the role of four proteins involved in establishing the axis. Each type of protein is represented in the model by a different colored bead, whereas corresponding transcripts are represented by small pipe cleaners of the same color. A molecular interaction of a protein binding a transcript to regulate its translation is modeled by students placing the appropriate colored bead on the appropriate colored pipe cleaner. In the model, any pipe cleaner (transcript) bound by a bead (protein) cannot be translated. For example, Bicoid binding to caudal transcripts in the anterior end of the embryo is modeled by placing a brown bead (Bicoid) on an aqua pipe cleaner (caudal transcript). When Bicoid is bound to caudal transcripts, translation is blocked. Thus, caudal protein is only produced in the posterior end of the embryo where there is no Bicoid (Figure 3A models the syncytial blastoderm before Bicoid/caudal transcript interaction, whereas Figure 3B models the embryo after the interaction and translation of caudal transcripts). Through this activity, posttranscriptional regulation becomes a tangible concept to students (Figure 4 and Figure 5).
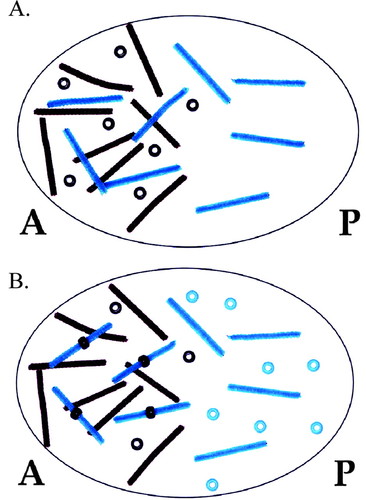
Figure 3. Demonstration of opposing Bicoid and Caudal gradients. (A) bicoid protein and transcripts are present in an anterior to posterior gradient, whereas caudal transcripts are present uniformly throughout the zygote. (B) Bicoid repression of caudal translation sets up a posterior to anterior gradient of caudal protein. Brown pipe cleaners represent bicoid transcripts; brown beads represent bicoid protein; aqua pipe cleaners represent caudal transcripts; aqua beads represent caudal protein. A, anterior; P, posterior.
Figure 4. Demonstration of the establishment of the Hunchback gradient. (A) Nanos represses maternal hunchback (mhunchback) translation in the posterior end of the embryo, whereas Bicoid activates expression of zygotic hunchback (zhunchback). (B) The combination of repression of hunchback translation in the posterior end and the translation of both maternal and zygotic hunchback transcripts in the anterior end generates an anterior to posterior gradient of Hunchback. Brown pipe cleaners and beads represent bicoid transcripts and protein, respectively; green pipe cleaners represent mnanos transcripts; green beads represent nanos protein; red pipe cleaners represent mhunchback transcripts; red pipe cleaners with a black stripe represent zhunchback transcripts; and red beads represent hunchback protein.
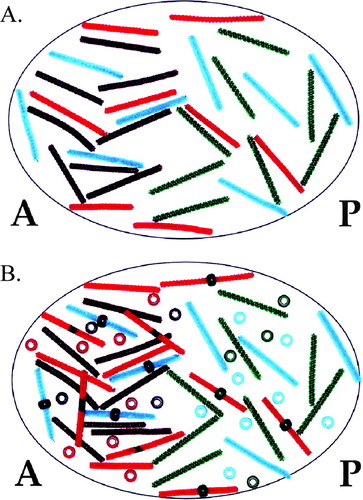
Figure 5. Modeling simultaneous molecular interactions involved in A/P patterning. (A) Maternal bicoid, nanos, hunchback, and caudal transcripts are distributed in their proper locations in the oocyte before fertilization. (B) Translational repression is indicated by beads placed on pipe cleaners. Gradients of Bicoid, Nanos, Caudal, and Hunchback have been established via these molecular interactions. Colors of pipe cleaners and beads are as described in Figures 3 and 4.
UNDERSTANDING MUTANT PHENOTYPES
Equipped with a clear understanding of how Bicoid, Nanos, Hunchback, and Caudal gradients are established, students are better able to predict how gene mutations will affect A/P patterning. While students are working with their models they are asked, “Suppose a nanos−/nanos− female (homozygous mutant at the nanos locus) was mated with a wild-type male. What would the progeny look like? Provide a CLEAR explanation for your answer.” Using the pipe cleaners and beads, students model this situation by removing the green pipe cleaners from the oocyte (Figure 6A). Once they begin to model translation of mRNAs, they realize there is nothing to repress hunchback translation in the posterior end of the embryo (Figure 6B). Nearly all students then recognize that there will be a shift in the Hunchback gradient toward the posterior end of the embryo. At the beginning of the activity, the role of Hunchback in A/P patterning was defined as repressing abdominal specific genes. Thus, students quickly realize that nanos mutants will possess abdominal defects. The power of this activity is that all students can visualize the effects of removing the green pipe cleaners. All students model a posterior shift in the Hunchback gradient. Most students are able to connect the gradient shift with an abdominal phenotype. Before the use of this activity, only the top tier students in the course could provide a clear explanation of mutant phenotypes.
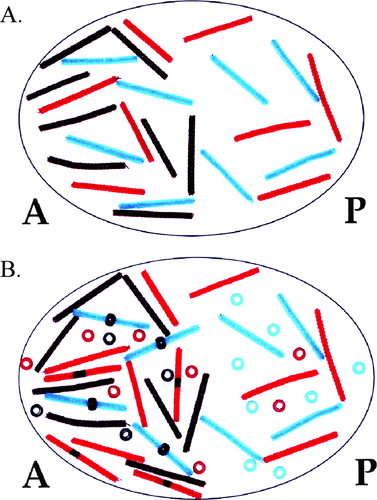
Figure 6. A posterior shift in the Hunchback gradient is observed in nanos mutants. (A) An oocyte lacking mnanos is modeled. Note the absence of green pipe cleaners compared with Figure 4A. (B) With no Nanos to repress translation of mhunchback, Hunchback protein is located in the posterior end of the embryo. Note the presence of red beads in the posterior end of the embryo compared with Figure 4B.
CAVEATS
This activity greatly simplifies the complex interactions required to pattern a Drosophila embryo. The model only includes four transcripts and four proteins. Additionally, the model does not explain how maternal transcripts are localized in the oocyte during oogenesis. It is important to note that this activity was developed for undergraduate students with little cell biology or biochemistry background. Thus, there is no distinction between direct and indirect molecular interactions involved in Drosophila A/P patterning. For example, Pumilio is not mentioned in the activity, even though Nanos interacts with Pumilio (which is bound to hunchback transcripts) to repress hunchback translation.
OUTCOMES AND ASSESSMENT
After working with the pipe cleaner and bead model, students have a firm understanding of how Bicoid, Nanos, Caudal, and Hunchback gradients are generated. They can visualize the process even without the aid of pipe cleaners and beads. By the end of the exercise, graphs like those represented in Figure 1 are comprehendible to students. Before working with the pipe cleaners and beads, students could see the concentration gradients on the graphs, but they did not understand how the gradients were generated.
Before students' initial work with the pipe cleaner/bead model, a pretest was completed assessing students' understanding of Drosophila A/P axis patterning based on self-review of the assigned textbook reading covering the topic. Knowledge survey type questions were included in the pretest to assess student confidence in their answers (Table 3; Nuhfer and Knipp, 2003). Student understanding of the same concepts were assessed in the unit exam, again including questions to assess their confidence in their answer. For three of the four pretest concepts assessed on the unit exam, there was a statistically significant difference in the percentage of students answering pretest questions correctly compared with the percentage of students answering unit exam questions correctly (Table 2). Additionally, there was a statistically significant difference in student confidence levels answering questions on unit exams compared with the pretest (Table 3). It is difficult to quantify the exact effect of the model on unit exam scores, because students received classroom instruction, practiced with the model, and studied independently in the time between the pretest and unit exam. However, informal assessments indicate the pipe cleaner/bead activity played a significant role in their understanding of A/P patterning in Drosophila as well as other topics covered in the course.
| Pretest question | Unit exam question | p value |
|---|---|---|
| Predict what an embryo lacking maternal bicoid transcripts will look like. | Embryos missing maternal bicoid transcripts will lack ______ structures. (Question from matching section of exam) (98%) | 0.003 |
| a. Lack anterior structures, but possess posterior structures | ||
| b. Lack posterior structures, but possess anterior structures | ||
| c. Fewer abdominal segments | ||
| d. More abdominal segments (27%) | ||
| Predict what an embryo lacking maternal hunchback transcripts will look like. | Embryos lacking maternal hunchback transcripts lack posterior structures. True or false (98%) | 0.002 |
| a. Lack anterior structures, but possess posterior structures | ||
| b. Lack posterior structures, but possess anterior structures | ||
| c. Fewer abdominal segments | ||
| d. More abdominal segments (15%) | ||
| Nanos protein is localized to the posterior end of the embryo, yet it contributes to an anterior to posterior gradient of Hunchback protein. How does this happen? | Nanos protein is localized to the posterior end of the embryo, yet it contributes to an anterior to posterior gradient of Hunchback protein. How does this happen? | 0.043 |
| a. Nanos activates hunchback transcription | a. Nanos activates hunchback transcription | |
| b. Nanos activates hunchback translation | b. Nanos activates hunchback translation | |
| c. Nanos activates hunchback transcription | c. Nanos represses hunchback transcription | |
| c. Nanos represses hunchback translation (22%) | d. Nanos represses hunchback translation (73%) | |
| When is anterior/posterior patterning in Drosophila initiated? | When is anterior/posterior patterning in Drosophila initiated? | 0.742 |
| a. In the developing oocyte | a. In the developing oocyte | |
| b. At fertilization | b. At fertilization | |
| c. As a syncytial blastoderm | c. As a syncytial blastoderm | |
| d. As a cellular blastoderm (29%) | d. As a cellular blastoderm (37%) |
| A. Please choose one of the following statements regarding your answer to question X.a | B.b | Pretest | Unit exam |
| a. I am fully confident of my answer. | |||
| Term 1 | 32 | 98 | |
| b. I am reasonably confident of my answer. | |||
| Term 2 | 42 | 98 | |
| c. I am unsure of my answer. | |||
| p = 0.007 | |||
| d. I do not have the knowledge base to answer this question. (i.e., This was a guess.) | |||
Six weeks after working with the pipe cleaner/bead model, students were asked to describe how the model helped them understand major concepts discussed in the Developmental Biology course. Representative student statements are listed below.
“This activity helped me understand what exactly is meant by a concentration gradient. The graphs of concentration gradients didn't mean much to me before the activity, but after I could picture how and where the concentrations increased and decreased.”
“This exercise, in my opinion, was absolutely wonderful … I definitely benefit from hands-on, visual representation of information. This way, when reading through my class notes, I am able to actually see myself putting colored pipe cleaners and beads in specific spots. This tangible object helps me so much with the somewhat abstract idea or thought.”
“It made it easier to visualize how changing one element would affect the rest of the gradients in the embryo. It helped me to understand that any change made during development can have drastic effects on the formation of the embryo. It helped me to see that proper development depends on a very delicate balance … one change can have dramatic effects. I even found myself visualizing this exercise during the exam.”
“This activity, more than anything, showed me that understanding the molecular interactions within the developing embryo takes more than just memorizing a bunch of molecule names: it taught me to keep in mind if something was a zygotic transcript or transcription factor, etc. and to use that to help me understand what each molecule is actually doing within a cell/embryo.”
“Since Drosophila was the first model system we studied, developing the A/P patterning model in the fly gave me a starting point from which to envision other gradients in other model systems, like in Xenopus.”
CONCLUSIONS
Student responses to this activity have been overwhelmingly positive, both while they are working with the model and throughout the course. After physically placing beads on pipe cleaners, students have a better idea of what a molecular interaction is and how it can affect the workings of a cell. Students no longer complain about the difficulty of the Drosophila unit of the course, and many students confess they actually had fun learning the material.
ACKNOWLEDGMENTS
I thank Dara Wegman-Geedey for insightful discussions regarding assessment and scholarship of teaching and learning work in the sciences. I am also grateful to my students who were willing to try something new and for suggestions on how to improve this activity.