The Design and Transformation of Biofundamentals: A Nonsurvey Introductory Evolutionary and Molecular Biology Course
Abstract
Many introductory biology courses amount to superficial surveys of disconnected topics. Often, foundational observations and the concepts derived from them and students’ ability to use these ideas appropriately are overlooked, leading to unrealistic expectations and unrecognized learning obstacles. The result can be a focus on memorization at the expense of the development of a meaningful framework within which to consider biological phenomena. About a decade ago, we began a reconsideration of what an introductory course should present to students and the skills they need to master. The original Web-based course’s design presaged many of the recommendations of the Vision and Change report; in particular, a focus on social evolutionary mechanisms, stochastic (evolutionary and molecular) processes, and core ideas (cellular continuity, evolutionary homology, molecular interactions, coupled chemical reactions, and molecular machines). Inspired by insights from the Chemistry, Life, the Universe & Everything general chemistry project, we transformed the original Web version into a (freely available) book with a more unified narrative flow and a set of formative assessments delivered through the beSocratic system. We outline how student responses to course materials are guiding future course modifications, in particular a more concerted effort at helping students to construct logical, empirically based arguments, explanations, and models.
INTRODUCTION
Biology is a complex subject, ranging from the molecular to the ecological and even to the emotional. It serves as the foundation for a wide range of careers, including healthcare and counseling, biotechnology, teaching, public outreach and policy, and academic research. It is also of immediate practical importance, informing a range of personal and public decisions from vaccination and reproductive rights to the desirability and dangers of genetic engineering. How biology is taught, and more importantly, understood by students, is therefore of increasing significance. Yet there is a common impression that introductory biology courses consist primarily of vocabulary lessons and often amount to superficial surveys of multiple topics without regard to their conceptual foundations, many of which emerge from physical or chemical principles. Whether students’ prior knowledge is sufficient to allow them to master and apply the materials presented seems rarely to be taken into account. Developing an understanding of underlying conceptual foundations, the observations upon which they are based, and the ability to practice their application takes time, a reality often overlooked in course design. The neglect of foundational understanding within an introductory course, under the aegis of coverage, can lead to persistent misconceptions and an inability to apply knowledge to new situations (Garvin-Doxas and Klymkowsky, 2008; Andrews et al., 2012; Couch et al., 2015). We have encountered this situation during the development of the Chemistry, Life, the Universe & Everything (CLUE) general chemistry project in the context of student understanding of the differences between covalent and noncovalent interactions (Williams et al., 2015) and their implications (Cooper et al., 2013; Underwood et al., 2016).
While it is unreasonable to overgeneralize about the details of the hundreds to thousands of college-level introductory biology courses taught in the United States, our own experiences with, and conversations about, such courses offered in molecular and cell biology–centered departments suggest that it is common to find that the complexities of evolutionary mechanisms are either not considered at all or treated superficially. In particular, social (kin and group) selection processes, which underlie the evolution and behavior of multicellular organisms and sexual reproduction, and a range of cooperative behaviors, from quorum sensing to altruism, are rarely discussed (Yarmolinsky, 1995; Crespi, 2001; Nowak, 2006). As an example, consider the different strategies used to deal with “social cheaters” within social and multicellular organisms (see Grosberg and Strathmann, 2007; McGinty et al., 2011). Similarly, stochastic processes, active at the population and the molecular and cellular levels, are largely overlooked. At the same time, it is often the case that students are asked to memorize facts, such as amino acid names and structures, reaction systems, components of signaling and metabolic pathways, without overt consideration of the subsequent utility of such facts. Students typically emerge from such courses without the ability to apply their knowledge productively to new situations.
In response to this situation, more than 10 years ago we set out to redesign a molecular and cell biology–focused introductory course to 1) include an extensive introduction to key evolutionary mechanisms, in part to address the fact that our current curriculum has no formal evolutionary biology component; 2) build out a coherent conceptual introduction to the molecular and cellular mechanisms involved in the origin, storage, and utilization of hereditary information; and 3) extend those concepts to social systems, including developmental and pathological processes. The result, Biofundamentals, originally relied on Web-based materials with no required textbook (Klymkowsky, 2007) and was taught using a semiflipped style (i.e., minimal lecture, in-class activities, and group discussions) and an in-class response system (clickers) to poll student thinking (Powell, 2003). A number of subsequent innovations, including social reading systems (the now defunct Highlighter.com and nota bene: http://nb.mit.edu), were introduced, together with formative assignments delivered through the beSocratic system (Bryfczynski et al., 2012a,b, 2015). As time passed, we gathered student data (see as examples Henson et al., 2012; Trujillo et al., 2012) and learned lessons from the development of CLUE. We used this evidence and experience to redesign Biofundamentals, transforming it from a website to a textbook and associated materials. While the materials are freely available, our efforts at refinement continue.
The Underlying Theory of Our Curriculum Design
It is well recognized that experts in a discipline have different knowledge structures compared with beginning learners. Experts’ knowledge is connected and contextualized into useful frameworks upon which new knowledge can be incorporated and applied to new situations (National Research Council [NRC], 2001; Schweingruber et al., 2012). In part, this reflects experts’ understanding of the underlying processes involved in the systems they are considering, which enables them to avoid an excessive cognitive load by ignoring irrelevant details and focusing on critical features and common, recurrent processes. In biology, these concepts include the accumulation of information based on mutation and selection; molecular interactions and their role in binding specificity and stability (dissociation); the thermodynamics of bounded, continuous, nonequilibrium systems (cells and organisms); the role of reaction coupling in the synthesis of nonstable molecules (nucleic acids, polypeptides, carbohydrates, lipids, etc.); the behavior of various molecular machines (motors, pumps, targeting, transport, repair, refolding, and degradation systems); and social interactions leading to cooperative behaviors, including multicellularity, sexual reproduction, and communal interactions.
The question then is how to help students recognize and focus on what is important, so that they can develop a deep and useful knowledge framework that can be applied throughout the tree of life to predict, understand, and explain biological phenomena. The traditional approach is, all too often, to teach a broad survey course in which students are introduced to a wide range of topics that may (or often may not) be returned to in later courses. These topics are typically not well integrated or connected; indeed, there is often an attempt to separate ideas into separate sections in a course (see Nehm et al., 2009). This is reflected in the recent publishing practice of offering instructors the ability to customize, that is, to assign sections of textbooks in their own preferred order, which suggests that the original textbook lacked a coherent organizational narrative. Presumably, the course (or curriculum) designer’s intent is that eventually the repetition of isolated ideas will coalesce and result in deeper and robust knowledge. Unfortunately, there is a great deal of evidence indicating that this is not how people learn complex subjects (NRC, 2001) and that even senior majors in biology-based disciplines have a fragmented knowledge base leading to ideas that are not scientifically normative. While there are reasons to believe that concept-type multiple-choice/true–false tests can overestimate the level of student understanding (Lee et al., 2011), their application suggests that, by the end of their undergraduate biology education, students’ understanding of key ideas is not at the level we might hope for or expect (see Klymkowsky et al., 2010; Couch et al., 2015). If we look at this process from the standpoint of how people learn, and how knowledge is constructed, we should not be surprised at this outcome.
Overarching Frameworks versus Chunks
There is general agreement that, rather than providing students with “chunks” of information, that is, topics presented without an overarching conceptual framework, learning should be structured around those core (recurrent and universal) ideas and skills that are important to a discipline (NRC, 2001). These core ideas and skills serve as the basis for students’ knowledge framework and can be extended if and when more sophisticated topics are encountered. There have been a number of national-level initiatives to identify the ideas central to biology as a discipline (e.g., American Association for the Advancement of Science [AAAS], 1993, 2001; NRC, 2003; Schweingruber et al., 2012). Vision and Change (AAAS, 2011) identified a small set of core ideas: evolution, structure and function, information-flow pathways, the transformations of energy and matter, and system behaviors—an analysis similar to one we had articulated (Klymkowsky, 2010). A parallel approach, the Advanced Placement redesign project, produced a similar list of ideas.1 It is important, however, to understand the difference between core ideas and topics.
Few would argue against the idea that evolution is THE overarching idea in biology (Dobzhansky, 1973; Jacob, 1982; Mayr, 1985; Bowler, 1989), but it is not a single topic. Modern evolutionary theory is a framework of ideas and observations, a mind-set that allows us to make sense of biological systems. In contrast to Darwin and Wallace’s original formulation (Darwin and Wallace, 1858), the modern theory of evolution rests on a number of core ideas, including 1) an understanding of how information accumulates in biological systems (the role of mutations, including gene duplications and deletions, and more recently, horizontal gene transfer and related topics); 2) an understanding of various mechanisms that include both adaptive and nonadaptive processes, including gene linkage; 3) sexual and social selection; and 4) the multigenic nature of most traits and the multifunctional roles of most genes. Without a clear understanding of these ideas, and the observations upon which they are based, it is difficult to make sense of much of what is taught in introductory courses. Yet it is entirely possible to present an introductory biology course without introducing or seriously considering the implications of evolutionary processes. As an example, consider the situation in which genetic drift is commonly presented to students. According to Price et al. (2014, p. 65), “Genetic drift is taught in major introductory biology textbooks … in the context of the Hardy-Weinberg equilibrium, a population genetics model that assumes that no evolution has occurred and, consequently, that the frequency of each allele remains constant,” a situation that must confuse students, since genetic drift does not occur under these conditions. Similarly, developing an understanding of how molecules interact with and dissociate from one another requires an appreciation of both the nature of intermolecular interactions and the processes that disrupt them, yet a survey of biology textbooks reveals that the mechanism of dissociation is (apparently) never explicitly discussed, while the factors that underlie the specificity and stability of molecular interactions (including entropic effects associated with the aqueous context of biological systems) are considered in a superficial manner. Not discussing these ideas explicitly makes developing an accurate understanding of reaction kinetics difficult, since it is the transfer of energy through collisions that (generally) supplies the activation energy needed for both reactions and dissociative processes. These are concepts key to understanding biological organization, from membranes to macromolecular complexes, and how such molecular machines produce the wide range of biological processes, from DNA error repair and protein folding to molecular syntheses, from growth and reproduction to sensory perception and movement. Finally, considering the behavior of multicellular organisms (such as people) is conceptually problematic if one does not address the social evolutionary mechanisms that led cells and organisms to cooperate in the first place (Trivers, 1971; Nedelcu et al., 2011).
While many introductory “cell and molecular” biology courses devote much time to the details of DNA replication, RNA transcription, and the translation of mRNA to produce polypeptides, there is generally little or no explicit consideration of what information represents, where information comes from, or how it flows within a system, which was the original point of Crick’s central dogma (Crick, 1970). That students reach the end of a molecular biology curriculum without a clear understanding of nonsense mutations (Couch et al., 2015) provides compelling evidence that many of these topics are poorly grasped, perhaps because the accumulation and modification of information is the direct result of evolutionary processes to which they have not been adequately introduced.
Students are often presented with the details of cellular anatomy, typically focused on eukaryotic cells, together with considerations of mitosis and meiosis, including the processes of chromosome segregation and recombination. Given the large amounts of disconnected information students are expected to remember, there is often little time to explore the foundational observations upon which these topics are based or how these ideas can be applied to new situations. Indeed, evidence that students understand the core ideas of the discipline can only come from assessments in which students learn how to use and apply their knowledge to new situations and are provided with useful feedback. At the same time, the narrative provided by evolution theory, which explains biological diversity, unity (homology), and complex social behaviors (including multicellularity and the endosymbiotic origin of eukaryotes) is often missing or poorly developed. As evidence Couch et al. (2015, p. 6) report that “While most students recognized that natural selection can cause changes in allele frequencies within a population … students had difficulty recognizing the potential impacts of other phenomena, such as founder effects, inbreeding, and new mutations.” At the same time, topics relating to social evolutionary processes (inclusive fitness, kin and group selection) involved in the development of multicellularity and social behaviors are absent. Similarly, in the course of the characterization of the Biology Concepts Instrument (BCI), we consistently found that students “apparently fail to grasp the ‘cost-benefit’ calculation implicit in evolutionary events” (Klymkowsky et al., 2010, p. 18).
DESIGN AND METHODS
The original version of Biofundamentals (MCDB 1111; Figure 1) was approved by the department and College of Arts and Sciences at the University of Colorado–Boulder (UC Boulder) as a four-credit course that replaced the existing versions of two courses: Introduction to Cell and Molecular Biology (MCDB 1150) and the accompanying laboratory (MCDB 1151). Subsequently, the course was taught as MCDB 1150–section 3, a three-credit course that met three times a week for 50 min. While we have not archived the course’s Web-page history, we have preserved the last Web version, which reflects the course’s original organization. Approximately one Web page was assigned for each course period.2 In the last four years (2011–2015) the Web-based materials were first adopted to be read by students (assigned randomly to groups of five to eight students each) using the interactive system developed by Highlighter.com. With the demise of Highlighter.com in 2013, students were assigned readings from the Biofundamentals text using the nota bene system (Zyto et al., 2012).3 As before, students were assigned at random into groups of five to eight students each. Beginning in 2012, students were also asked to complete a beSocratic activity before each class (the system was running on servers at Michigan State University). Both activities were mandatory for students in the class, with compliance influencing students’ final grades. The use of deidentified beSocratic data and nota bene data were judged exempt by the UC Boulder Institutional Review Board (IRB 0304.09 and 15–0347). Student responses to beSocratic activities were characterized based on predefined rubrics to identify apparent issues in understanding underlying ideas. In addition, questions raised during in-class discussions were noted and used to inform subsequent revisions of first the website and then the text and associated beSocratic activities. Similarly, exam questions were analyzed using a rubric (three-dimensional learning assessment protocol [3D]-LAP) generated in the course of American Association of Universities (AAU)-funded studies at Michigan State University to characterize the nature of exam questions on the basis of a number of criteria, including whether the answers involved disciplinary core ideas and cross-cutting concepts and practices (Laverty et al., 2016); these rubrics are illustrated for the final exam given in 2015 (see Supplemental Material 3). The course was routinely taught with the aid of learning assistants (Otero, 2006), undergraduate students who had previously taken the course, who were taking a concurrent course in science pedagogy, and who met weekly with the course instructor to discuss students’ issues.
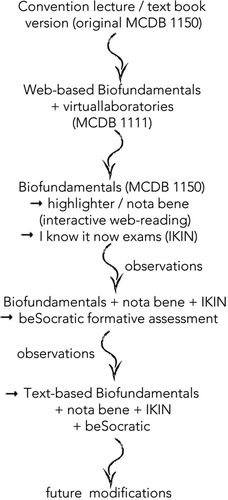
FIGURE 1. Schematic of the design progression from a conventional introductory molecular biology course to Biofundamentals, which has a largely flipped and interactive course design focused on a set of core concepts. At various steps in the process, observations from student interactions with course materials led to revisions in the representation of materials.
PROCESS DESCRIPTION AND OUTCOMES
In 2005, driven in part by students’ responses to open-ended questions used in the process of the development of the BCI and its subsequent administration (Garvin-Doxas and Klymkowsky, 2008; Klymkowsky and Garvin-Doxas, 2008; Klymkowsky et al., 2010) and our (M.W.K.) own experiences teaching the existing version of the introductory course, we redesigned the introductory course for MCDB majors at UC Boulder. While this redesign effort predated the Vision and Change report, it was organized around a similar set of core ideas (as discussed earlier). Here, we describe the “lecture” aspect of the course. Since its inception, the course has been taught in a scale-up type classroom (capacity between 100 and 120 students; Beichner et al., 2000), eventually using an Apple TV/iPad–based wireless interactive system that allows the instructor to move freely among the students and to share, for in-class discussion, student work and responses. As an example, student drawings (typically generated through group activities) were photographed using the iPad’s camera and projected for all students to consider.
Biofundamentals Course Narrative
Since its inception, we anchored the course in the three central observations in biology: 1) the diversity (multimillions) of distinct organisms; 2) the ability to organize all organisms (both extinct and extant) into a hierarchical (Linnaean) branching tree diagram that reflects ancestor–descendant relationships; and 3) the underlying structural unity (homologies) between all organisms, which includes the presence of glycerol lipid–based membranes, common chemical reaction networks, the use of double-stranded DNA and a common genetic code, and an mRNA/tRNA/rRNA-based translation system, as evidence for a common ancestor.
The story line of the course (Figure 2) begins 1) with how we study living systems scientifically. This leads to a consideration of the continuous nature of living systems (cell theory) stretching back over billions of years to the last common ancestor, and 2) the evolutionary mechanisms that shaped the structures and behaviors found in living systems. Together, these processes generate the diversity of species and provide an explanation for their common features. The course then moves to consider the physicochemical properties of living systems, including 3) molecular interactions with water (entropic factors) and the capture of energy to drive these continuous nonequilibrium systems (organisms). We then consider 4) the nature of the molecular machines (including information storage devices [DNA and RNA], ribosomes, molecular motors and pumps, chaperones, and proteasomes)—their structure, assembly, regulations, and functions. In the final section, 5) higher-order behaviors are considered, involving regulatory networks leading to cell division, organismal life cycles, and intercellular interactions. There are a number of reasons to begin this way, rather than with molecule-level descriptions that many biology students are ill prepared to understand. Most obviously, evolutionary relationships (homologies) provide a framework within which to present the common structures and processes found in biological systems.
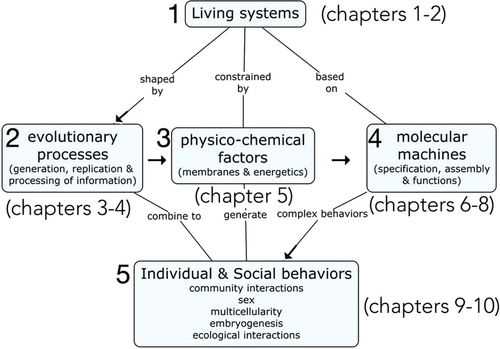
FIGURE 2. Diagram of the course narrative for Biofundamentals. The flow of the course (from 1 to 5) is indicated by arrows, with recurrent concepts linking topics indicated by lines and words.
The Place of Chemical Concepts in Biofundamentals
Many cell and molecular biology courses begin with molecular-level processes that involve understanding macromolecular structures and interactions for seemingly good reasons. Indeed, an understanding of biological systems rests on the physicochemical properties of matter, the thermodynamics of chemical reactions systems, the historical nature of systems that have been “running” uninterruptedly for more than ∼3.8 billion years, and the impacts of stochastic and selective processes on biological diversification and adaptation. In fact, chemistry is often called out as a prerequisite in biology textbooks, and presumably the courses that use them, but our own work on the design and evaluation of a general chemistry course suggests that, after taking general chemistry, students often emerge without a working understanding of many core concepts (see Cooper et al., 2010, 2012; Williams et al., 2015). Moreover, if students are concurrently taking a general chemistry course, the pacing and sequence of such a course means that the materials that a (molecular and cell) biologist might see as prerequisites are often not considered until the second semester, and then not often mastered (Underwood et al., 2016). For example, to understand the structural and functional significance of hydrophobic versus hydrophilic amino acid side chains (in the context of polypeptide and protein structures), it is necessary to appreciate the interplay between enthalpic and entropic factors that influence both inter- and intramolecular interactions. Similarly, understanding the significance of weakly acidic and basic amino acid side chains within a protein structure implies that students grasp how environmental pH influences whether these moieties are charged or not, as well as how charged groups interact with water, dissolved ions, and one another. More to the point, there is simply no compelling evidence that students understand the significance of the structural properties of the complex macromolecules to which they are being introduced, whether lipids forming a membrane or the folding of a multisubunit protein complex (e.g., see Williams et al., 2015). This means that students cannot reason through what they are being taught and are actually forced into rote memorization of the processes and ideas with which they are presented. The result is knowledge that is not contextualized or linked to observations; knowledge that is difficult to integrate into a framework that is meaningful to the student.
The theory of meaningful learning (Ausubel et al., 1978; Mintzes et al., 1998; Novak, 2002) posits that, for students to learn in a way that allows them to use their knowledge, what they learn must be linked to what they already know, that is, their prior knowledge. Just as importantly, it must be clear to the student what the purpose (i.e., usefulness) of that knowledge is. If what we expect students to learn is disconnected from either past or future knowledge, it is unlikely they will be able to appreciate its relevance or to use it appropriately in new scenarios. In this light, beginning at the molecular level can seem abstract, making it difficult for students to understand the purpose of what they are being asked to learn. In the absence of an appreciation of biological phenomena, such as species diversity, structural and molecular homologies, and the effects of mutations on phenotypic variation, it is unlikely that students can make sense of, and connections involving, what they are learning. In contrast, once biological continuity and evolutionary concepts have been introduced (and used in various contexts), we can ask students how various molecular processes can be modified in different systems to produce different outcomes. These are skills that can enable students to consider how the effects of mutations on the development of new molecular functions interact with and depend upon processes such as gene duplication (see Bergthorsson et al., 2007) and recombination.
Learning Outcomes and Their Assessment (Exams)
An important element of any course’s design is to have a clear vision (and articulation) of the desired learning outcomes and then to assess those outcomes with tasks that elicit evidence that the students have indeed learned and can use the knowledge they have been taught. In our case, the learning outcomes center around developing an appreciation of (i.e., the ability to recognize the relevance of) and the ability to apply evolutionary mechanisms, physicochemical principles, and network behaviors to a particular biological situation, together with an understanding of those elements of biological systems that cannot be deduced from first principles, such as the direction of nucleic acid polymerization, the use of double-stranded DNA as the genetic material, the common organizational features of cells, and so forth. We have set out course goals in a one-page document, supplied as Supplemental Material 1 and available online to students. A central aspect of these goals involves developing in students the skill of being able to recognize (and articulate) the ideas their answers are based on. The grading rubrics applied to exam answers (shown in Supplemental Material 3) illustrate what we mean when we claim to be encouraging students to construct explanations and arguments. That is, they must articulate more than the facts and must support their answers with scientific principles and reasoning.
As a general rule, courses can focus primarily on maximizing student learning or the sorting of students. The use of curves in grading explicitly acknowledges this dichotomy. “Curve-grading forces students to compete with each other, whether they want to or not” (Seymour and Hewitt, 1997; see also Humphreys et al., 1982; Covington, 1992)4 and can “dissociate grades from any meaning in terms of content knowledge and learning” (Schinske and Tanner, 2014, p. 118). In addition, there are often complicating factors in play, including the correlation between students’ ranking of a course and its instructor and students’ expectations with respect to their grades (see Boring et al., 2016) that can, often unconsciously, impact the construction and grading of exams.
We have made an explicit attempt to emphasize the importance of learning over sorting through what we term “I know it now” (IKIN) exams. Students take three midterm exams. At the time of the final exam, they are offered the opportunity to take one, two, or three IKIN exams, each associated with a particular midterm (see Supplemental Material 3). IKIN exams are typically worth up to 20 points (midterm exams are worth 100 points). IKIN exams are optional, but if a student selects to take one, his or her score on the IKIN exam is added to the associated midterm exam (midterm + IKIN) up to a maximum score of 100 points for any midterm exam. Throughout the semester, students are repeatedly reminded that their midterm scores remain tentative until they have completed the final exam and those IKIN exams they decide to take. Because course grades are not curved, points scored through IKIN exams do not negatively impact other students’ grades (unless one argues that a grade’s value is based on its exclusivity [scarcity] rather than as a measure of learning).
Collecting Evidence for the Transformation of Biofundamentals
One of the advantages of developing a course de novo (rather than following an existing text) is that it is possible to use a design-based research approach (Brown, 1992) to improve the course over time based on evidence from a range of sources. While the most obvious sources of evidence are the formative and summative assessments used in a course, recent technological advances have made others available. For example, we used social reading applications (initially Highlighter.com and later nota bene; Zyto et al., 2012),5 so that students (assigned to small groups) can comment on the text and respond to one another (Figure 3). We used the system to encourage reading of relevant material for class (10% of the class grade was assigned to student reading and responses to other students within the system) and to generate questions and conversations that can be continued and resolved in class. It also enabled us to identify parts of the websites and later the text that were confusing for students. As examples, responses to the section on genetic drift suggested that (at least some) students understood its implications (see Figure 3), while responses to materials on chemical reactions and their rates and student thinking about molecular interaction networks (Trujillo et al., 2012) and the link between gene and phenotype (Henson et al., 2012) indicated a need to reframe materials.
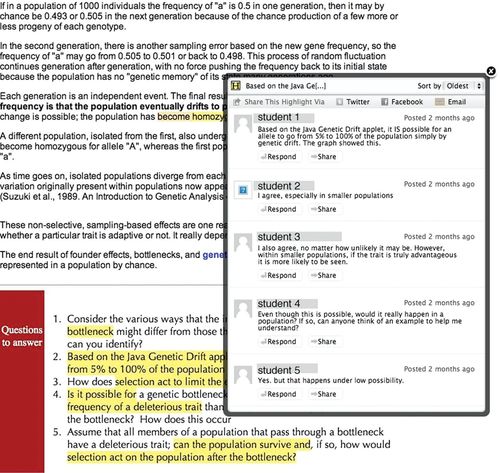
FIGURE 3. To illustrate the use of the Highlighter system, we show a page from the Biofundamentals website circa 2012; each yellow highlight indicates a student comment/conversation. One of these conversations (box) is expanded and reveals student thinking about the probability of allele loss due to drift and the influence of selection. These topics are extended in class.
Designing Assessments as Sources of Evidence
A key element of any course is how learning is assessed. Inauthentic tests lead to ersatz (i.e., “an artificial and inferior substitute or imitation,” per the Merriam–Webster Dictionary) learning (McClymer and Knowles, 1992). Indeed, while course grades are often (by necessity) used as a proxy for learning, this is not necessarily justified. In addition, we need to think carefully about the messages that assessments (tests, quizzes, and homework assignments) send to students. While it is clearly important that students “know” things (i.e., facts, such as that nucleic acid polymerization in biological systems proceeds in a 5′ to 3′ direction), it is the connection between these facts and ideas that grows into understanding. It is tempting, particularly when pressed for time, to develop simple assessment items that test facts. Many traditional assessments, particularly in larger introductory courses, are “forced-choice” in format, that is “multiple-choice tests, fill-in-the-blanks, true-false, matching, and the like.”6 It has been found that forced-choice formats overestimate the level of student understanding and lead to an emphasis on memorization and/or recognition (Lee et al., 2011). As an aside, the same can be said for forced-choice concept tests; although they are useful in revealing what students find difficult, they do not have the resolution to reveal what students actually know. While it is the case that more informative formats (open response) are more “expensive” to analyze, they are necessary if serious conclusions about student learning are to be drawn. The situation is analogous to that found in many scientific studies, in which more expensive and time-consuming analyses (e.g., the use of real-time polymerase chain reaction [PCR] vs. standard PCR) are often required to make quantitatively meaningful statements. That said, it is clear that students need explicit training on how to construct scientific explanations and arguments, based on the use of empirical data and the application of explanatory concepts (Cooper, 2015). On the basis of an analysis of student responses, our intention is to increase the time devoted to in-class analysis of student explanations, in part by giving students a copy of a past first midterm exam at the start of the course and using those questions as in-class exemplars about what an adequate response contains.
Exam Questions
The point of reorganizing a curriculum around core ideas and skills, including the ability to deconstruct a particular scenario into its components and to apply unifying (and relevant) concepts, including graphical representations, is that the questions we ask to assess student understanding are directly connected to one or more concepts. The questions we use typically have a two-part structure: either a multiple choice followed by an explanation or a justification for why a correct response is correct or an incorrect response is incorrect. Alternatively, students are asked to generate a graph or diagram of a particular process and then supply a justification or extrapolation of the effects of a perturbation (Supplemental Material 3). We also give students the option to select “no idea,” a choice that is awarded 1 out of 5 possible points (per question) and designed to allow students to avoid wasting time on questions they do not feel they understand (Klymkowsky et al., 2006). Questions are graded using a predetermined rubric established through conversations between instructors, learning assistants, and graduate teaching assistants.
Using the recently developed 3D-LAP rubric, we have been able to look back on past exam questions to identify those that require students to display their knowledge of information and the ways that knowledge can be used. As an example of what (in retrospect) we would consider a weak question, consider the question in Figure 4A, which asks students to recognize a definition, and so is primarily a memorization question—it does not contain a core idea, an explanation, or an application. In contrast, we would consider the questions in Figure 4, B and C, to be significantly stronger, because they involve understanding a core idea (how structural properties produce physicochemical properties and an explanation that provides a cause and effect). If answered appropriately, this question elicits evidence about how students understand 1) how bond polarities affect molecular polarity, 2) how molecular polarity affects interactions between molecules, and 3) how interactions between molecules affect phase-change temperatures. Similarly, the question in Figure 4D requires an understanding of 1) how reaction coupling is involved in driving thermodynamically unfavorable processes (i.e., the packing of DNA into a small volume) and 2) how proteins associate with DNA to overcome DNA’s net negative charge. It involves the processes of energy transfer and structural interactions that alter molecular properties (repulsion) and it involves an explanation for why the process is unfavorable. We believe it is the associated explanations that are the important aspect of these assessment tasks. They provide direct evidence about what it is that students know and are able to do; in subsequent papers, we will report on our analysis of student responses from these types of assessment tasks.

FIGURE 4. Four example questions that illustrate the range from definition to explanation.
beSocratic Formative Assessments
With the development of the National Science Foundation (NSF)-supported beSocratic graphics-based formative assessment system (Bryfczynski et al., 2012a,b, 2015), we were able to introduce Web-based formative assessments, initially as a supplement to in-class clicker questions and later replacing them. Students were asked to complete assigned beSocratic activities before class. While their responses were not graded, students received participation points for good faith completion of the activities.
Observations from beSocratic assessments influenced the course in a number of ways. First, analysis of student responses revealed which particular concepts or aspects of their application were poorly grasped by a significant fraction of students. This led to reconsidering how those concepts were presented in the text and through in-class activities. One immediate result was to add new “questions to answer/questions to ponder” into the text, questions that are often used in class as the focus of activities and discussions. Second, as we added more beSocratic exercises that call upon students to plot data and generate and interpret graphs; we have found that many students have difficulty analyzing and interpreting data. While we did not add a section on plotting data to the text, we did increase our emphasis on this skill through in-class activities by repeatedly asking students to articulate how axes were labeled and what particular graphs implied about the behavior of the system under consideration.
Similarly, as we introduced beSocratic activities that considered stochastic processes, a problem that we had suspected but had seriously underestimated emerged; in addition to having difficulty generating graphs describing the behaviors of large populations, few students correctly described the behavior of small populations. This has led us to increase our current and future emphasis on 1) how to draw graphs based on data, 2) how to predict behaviors and draw conclusions based on data presented through graphs, and, from the perspective of stochastic processes, 3) how large populations differ in their behavior from small populations (Klymkowsky et al., 2016). As part of this focus, we have included materials based on the work of Vilar et al. (2003) and, more recently, Uphoff et al. (2016) on the roles of regulatory noise, including the stochastic processes that underlie DNA–protein interactions (Elowitz et al., 2002; Maheshri and O’Shea, 2007). We are in the process of determining whether these changes improve students understanding or whether further design changes (including more time on task) are needed.
In many cases, the presentation of materials in the text was considered clear (based on part on students’ Highlighter.com and nota bene comments); nevertheless, student responses revealed deficiencies that needed to be addressed. This is an example similar to that learned from lecture-only classes (Powell, 2003), namely, that simply telling someone something, no matter how clearly, does not mean he or she will learn it, that is, appreciate its implications, or be able to apply those ideas to a particular situation. In the activity illustrated in Figure 5, it was clear that most students failed recognize that the gradients formed in biological system are, by their nature, limiting; as a proton-based electrochemical gradient forms, the movement of more protons is opposed by presence of the gradient, more and more energy is required to move H+s against the electrochemical gradient (an idea addressed in a question from chapter 5: “What would limit the ‘size’ of the H+ gradient that bacteriorhodopsin could produce?”). This led us to stress the behavior of ions (and other molecules) and how gradients influence their net (rather than individual) movements (flux), and in class we directly consider the factors that limit net flux.
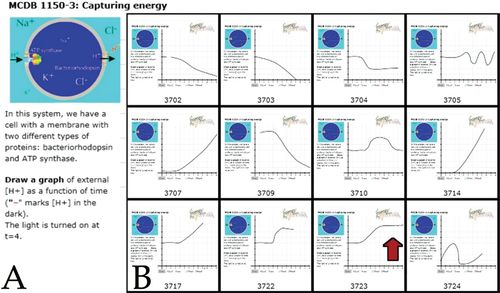
FIGURE 5. Student’s graphical responses captured through the beSocratic system reveal that many do not generate graphs that reflect a correct analysis of the problem. While a number of students indicate that external [H+] begins to increase after the light is turned on, only one (red arrow) indicates that the increase in [H+] levels off as the system reaches a steady state. Examples from student work are used as the basis of in-class group activities.
In a second example, many students have difficulty identifying all of the polarized bonds within a molecule (Figure 6) and are often confused when asked to distinguish such bonds from hydrogen bonds (see Williams et al., 2015). The ability to identify polarized bonds is central to an analysis of the molecular factors driving nucleic acid and protein structure and membrane formation. While mentioned in the text, it is clear that students need explicit (and repeated) practice with this idea: we therefore build on beSocratic activities through in-class activities. For example, we explicitly ask students to consider and discuss amino acid side chains in class in order to identify polar and nonpolar bonds, so students can better recognize (predict) the properties of the different amino acid side chains (rather than being asked to memorize them).
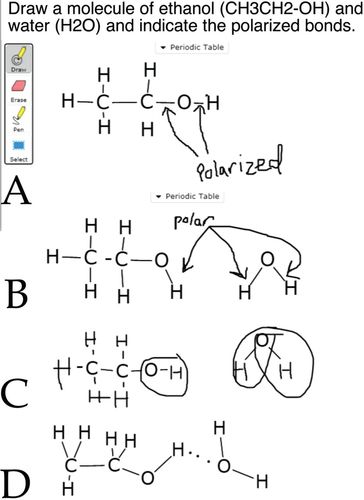
FIGURE 6. When asked to identify the polarized bonds within a simple molecule, such as ethanol (A), students often fail to recognize that both the C-O and O-H bonds are polarized (B and C), while some students appear to confuse H bonds between molecules with polarized covalent bonds (D). These responses are used in the context of in-class considerations of the hydrophilic/hydrophobic features of nucleotides and amino acids.
At the same time, the responses of students to beSocratic assessments can indicate the need to reconsider how particular ideas are presented both in the text and in class. To illustrate this, we present notes from the evaluation of student responses (carried out by J.D.R.) to a question posed during teaching of the Web-based (2012 academic year) version of the course (Supplemental Material 2). The distinction between evolutionary ancestry (homology) and convergent evolution (homology and analogy) is one that can be confusing to students (there seems to be little published on this topic, but we noted such confusions in the development of the BCI; K. Garvin-Doxas and M.W.K., unpublished observations). Biological structures, processes, and traits (including genes and proteins) are homologous if they are derived by ancestry from a structure, process, or trait present in the common ancestor of the organisms being compared. They are either homologous or not; they cannot be partially homologous. Differences arise during their subsequent, independent evolutionary histories. Analogous structures, processes, and traits are those that arose independently, that is, they were not present in the organisms’ common ancestor. They represent independent solutions to what are often similar evolutionary situations or opportunities, such as the benefits of flight or sight; the presence of a new energy source; a particular ecological niche; or the presence of toxins, predators, or pathogens.
To gauge how well students understand the idea of homology, we asked, “What type of evidence would convince you that structures in two different organisms are homologous?” Two major points in a student’s response were taken as a reasonable proxy for an accurate understanding: 1) Does the student mention that the structures compared are similar in detail? 2) Does the student mention the need for evidence that the structures were present in the common ancestor of the organisms being compared? Nearly all students (∼91%, or 87/96) indicated the need for structural similarity, but only ∼46% (44 of 96) of students indicated that the structures needed to be related by descent to be homologous. In terms of actual reliance on empirical evidence, however, less than 20% (16/96) of students indicated the need for either genetic or fossil record data in order to conclude common ancestry. Here are two such statements:
“I would like to go back into the fossil record to see if these two organisms had an identifiable common ancestor, where perhaps these two similar structures came from.”
“DNA evidence would convince me that structures in two different organisms are homologous. This is because with mere cosmetic data, it is difficult to determine unambiguously whether the traits are similar because of convergent evolution or because of common ancestry. If I had an unusual boney ridge serving some purpose in a bird which appeared similar to such a ridge in another bird, I would not know if they were homologous or analogous without DNA evidence.”
Observations such as these have led us to introduce, and to increase our emphasis on, the importance of relying on evidence at the various points in the course at which considerations of homology are relevant and considering whether these changes improve understanding or whether further design changes (including more time on task) are needed. For example, increasing our emphasis on the importance of specifically calling out the observations or principles upon which an argument is being made; for example, why a correct answer is correct or why an incorrect choice is wrong.
Moving from the Web to Text
The Web-based version of the course was taught for 8 years, with more than 900 students participating. During that time, two of the authors (M.W.K. and M.M.C.) initiated an NSF-funded collaboration to design a transformed general chemistry course, Chemistry, Life, the Universe & Everything, or CLUE (Cooper and Klymkowsky, 2013) and the development of the beSocratic system (Bryfczynski et al., 2015). CLUE was based on an underlying narrative, the development of chemistry and its relevance to living systems as a framework for introducing the core ideas of bonding and molecular interactions; structure and properties; and energy, change, and stability. The dialogues between a cell and molecular biologist and a chemist and chemical education researcher resulted in a productive process that challenged many often unrecognized assumptions driving conventional chemistry course design and resulted in a course that produced measurable improvements in students’ understanding of key ideas (and reduced drop/fail/withdraw rates). Longitudinal studies of matched cohorts of students from CLUE and traditional courses showed that the differences between these groups persisted all the way through a subsequent year of organic chemistry (Williams et al., 2015; Underwood et al., 2016). In the case of Biofundamentals, although there was a preexisting Web-based course, the process of generating a textbook benefited from many of the same types of synergistic interactions but with roles reversed. That is, does the chemist understand why a particular observation or concept is introduced, and in what order? Can the biologist defend the treatment of a particular topic as being essential to the course? In subsequent revisions, we had critical input from an evolutionary biologist (E.B.). Throughout the course design process, the distinctions and similarities between thinking in physics, chemistry, and biology was reinforced by our experiences in the NEXUS physics project (Redish et al., 2014). It is our conclusion that much can be gained in terms of clarity from such a collaborative approach to course design.
While the topics presented in the Biofundamentals book available online at http://tinyurl.com/nvfo97h and http://bio.libretexts.org/TextMaps/Map%3A_Biofundamentals_(Klymkowsky_and_Cooper) are similar to those present in the Web version (Figure 1), there were a number of changes necessitated by the distinctly different narrative flow between the two. In particular, we take information from online comments by students (Figure 3), analysis of past exams (Figure 4), and responses to beSocratic activities (Figures 5 and 6) to concentrate on ideas that students find problematic. This has led to an increased emphasis on drawing, reading, and interpreting the implications of graphs and the expansion of certain themes, including biological economics (cost–benefit analyses), social evolutionary mechanisms, and the impact of genetic drift and other nonadaptive processes, such as gene linkage (using vitamin C dependence in primates as an example). The role of stochastic processes, like genetic drift, are reinforced through consideration of molecular-level processes, including molecular movements, reaction kinetics, and the noisiness of gene expression and molecular degradation (Elowitz et al., 2002; Vilar et al., 2003; Maheshri and O’Shea, 2007; Pedraza and Paulsson, 2008; Uphoff et al., 2016). More emphasis has been given to the general concept of information, for example, when considering the factors that influence the location of a protein within a cell (signals and receptors) and the explicit recognition that cellular topology is, like genetic information, passed on to offspring during cell division. In addition to reconsidering our overall approach to the presentation of reactions, we also consistently referred to reactions as thermodynamically favorable (rather than spontaneous) or unfavorable, together with factors involved in reaction coupling and reaction rates. As a minor point, we referred to hydrogen bonds as “H-bond type electrostatic interactions” to emphasize how they differ from covalent bonds. We have also become more explicit about asking students what it takes to conclude that traits, structures, and processes are homologous. We have attempted to more explicitly acknowledge the counterintuitive nature of many biological processes, for example, having students consider the number of unproductive to productive collisions associated with polypeptide synthesis. We also more directly and repeatedly ask students to consider how the molecular machines involved in DNA damage repair, protein assembly (chaperones), and such can be expected to act, including being explicit about their dependence on coupled, thermodynamically favorable reactions and molecular interactions.
CONCLUSIONS
We have presented the history and rationale for the design and the ongoing transformation of Biofundamentals. The course’s design rests on established theories of learning that stress the importance of focusing on a scaffolded progression of core ideas and their application. Both the highly collaborative process of writing and revising the text and the overall story line of the course helped make these ideas explicit, beginning with macroscopic phenomena and then delving into molecular-level mechanisms. We use a student data–driven design approach based on observations from previous versions of the course to identify challenging ideas and skills, leading to changes in course emphases in terms of extended discussion and time on task of difficult subjects. The result is a course that remains focused on core ideas and observations and their application to new biological phenomena, with the recognition and removal of more peripheral materials (e.g., the names of lipids, the memorization of amino acid structures) as necessary. The subsequent evolution of the course will continue to be driven through modifications of beSocratic and in-class activities, together with changes in the text in response to student questions and conversations.
FOOTNOTES
1 https://apstudent.collegeboard.org/apcourse/ap-biology/course-details.
2 http://virtuallaboratory.colorado.edu/Biofundamentals-web.
3 http://ccnmtl.columbia.edu/enhanced/solutions/mits_nb_collaborative_reading_done_right.html.
4 http://insights.wired.com/profiles/blogs/breaking-the-curve-why-the-bell-curve-is-hurting-our-college#axzz40TRO5Q89;http://deltacollegian.net/2014/03/23/curve-grading-bad-students.
5 https://oeit.mit.edu/gallery/projects/nb-pdf-annotation-tool.
ACKNOWLEDGMENTS
The development of the original version of Biofundamentals (MCDB 1111) was carried out as part of a sabbatical project and was made possible through generous gifts from Spencer I. Browne and Hillary L. Browne. Data from the BCI project (NSF 0405007) were used to inform the original design of the course. The development of the beSocratic system was funded through NSF projects DUE 1341987and DUE 1043707. We have benefited from the involvement and insights of a number of colleagues, particularly Tristan McClure-Begley and Jennifer Martin; learning assistants; and graduate teaching assistants, particularly Joe Rokicki; and from the work of Natalie Jones, who designed the original beSocratic activity on genetic drift.



