Teaching Metabolism in Upper-Division Undergraduate Biochemistry Courses using Online Computational Systems and Dynamical Models Improves Student Performance
Abstract
Understanding metabolic function requires knowledge of the dynamics, interdependence, and regulation of metabolic networks. However, multiple professional societies have recognized that most undergraduate biochemistry students acquire only a surface-level understanding of metabolism. We hypothesized that guiding students through interactive computer simulations of metabolic systems would increase their ability to recognize how individual interactions between components affect the behavior of a system under different conditions. The computer simulations were designed with an interactive activity (i.e., module) that used the predict–observe–explain model of instruction to guide students through a process in which they iteratively predict outcomes, test their predictions, modify the interactions of the system, and then retest the outcomes. We found that biochemistry students using modules performed better on metabolism questions compared with students who did not use the modules. The average learning gain was 8% with modules and 0% without modules, a small to medium effect size. We also confirmed that the modules did not create or reinforce a gender bias. Our modules provide instructors with a dynamic, systems-driven approach to help students learn about metabolic regulation and equip students with important cognitive skills, such as interpreting and analyzing simulation results, and technical skills, such as building and simulating computer-based models.
INTRODUCTION
To ensure that the United States continues to be globally competitive in science, technology, engineering, and math (STEM), students entering the workforce must be adequately prepared to meet emerging challenges. As evidenced by various calls to action, the education community is working to address this need (American Association for the Advancement of Science [AAAS], 2011; National Research Council [NRC], 2012, 2013). These calls have prompted many educators to re-evaluate the ways in which they approach science education and find ways to identify and develop innovative and evidence-based solutions to educational problems (White et al., 2013; Howell et al., 2018, 2019; Kramer et al., 2018). One of the shifts in life sciences education has been to incorporate computer-based models to enhance students’ cognitive skills, such as the ability to reason quantitatively, as well as technical skills, such as the ability to use models to support thinking and problem solving (National Science and Technology Council, 2018). Moreover, the ability to analyze and interpret data using appropriate modeling and simulation tools has been broadly established as a core competency for undergraduate students (AAAS, 2011; NRC, 2013). The field of biochemistry is no exception, and the American Society for Biochemistry and Molecular Biology (ASBMB) and others have detailed the need for improving students’ foundational knowledge and skills in these areas (Tansey et al., 2013).
In biology and biochemistry, knowledge of metabolic networks is fundamental but also conceptually challenging. Students must understand the structure and regulatory relationships of metabolic networks to explain various phenomena that are characteristic of normal and pathological conditions. Students must also understand that regulatory relationships are important to maintain homeostasis. The ASBMB has identified five core concepts in biochemistry that all students should know upon graduation, including the concept of “homeostasis” (Tansey et al., 2013). Tightly tied to these core concepts are five threshold concepts that demonstrate a foundational understanding in a particular discipline (Loertscher et al., 2014). “Biochemical pathway dynamics and regulation” is one of these threshold concepts (Loertscher et al., 2014). To master metabolism, students must understand these fundamental concepts, the interrelationship between concepts, and additional “linking ideas” that underlie the interrelationships (Waheed and Lucas, 1992). Despite repeated exposure to the same biological system, students face difficulties when learning about biochemical pathways that can ultimately lead to the persistence of misconceptions about metabolism (Anderson et al., 1990; Michael et al., 1999; Wilson et al., 2006; Brown and Schwartz, 2009; Dauer et al., 2014). Schultz (2005) highlighted the “learning demand” on students who study metabolic pathways as follows: 1) knowing the particular chemical transformation involved, 2) evaluating the thermodynamics of each step, and 3) comprehending the biological context. As a result, the amount of information contained in a single pathway of a metabolic network can quickly overwhelm students, making it difficult to interpret the function and regulation of interconnected networks and how they maintain the function of the organism (Anderson et al., 1990; Hmelo-Silver and Pfeffer, 2004; Schultz, 2005; Brown and Schwartz, 2009).
Achieving a deep conceptual understanding of how metabolic networks function also relies on appreciating their dynamic nature. However, learning about the dynamics of a system through the use of static images and descriptions could be difficult for students. Most biochemistry textbooks focus on the details of individual enzymatic steps of metabolic pathways, and typical textbook readings only broadly discuss the integration and dynamics of metabolic pathways. Therefore, textbook readings and images may not allow students to fully appreciate and experience the dynamics of the metabolic networks they are studying.
Students’ struggles with metabolic systems may also be partly attributed to their inability to understand or predict how a system will respond to changes (i.e., to adopt a systems-thinking perspective). The systems-thinking perspective does not solely rely on conceptual knowledge of the system or appreciating its dynamic nature. Instead, it requires an analytical approach (Arnold and Wade, 2015; Verhoeff et al., 2018). In a systems-thinking approach, critical thinking and problem-solving are needed to explicitly explain how specific components interact and how these interactions allow the system to respond dynamically to changing environments. When students adopt a systems-thinking perspective, they must therefore conceptualize systems as interconnected processes that are themselves nested within larger systems and whose functions and responses can be understood and explained based on the interactions between the components (i.e., the mechanism; Abrams and Southerland, 2001; Wright et al., 2014).
The use of model-based learning encourages students to make their thought processes explicit, and this approach to learning can increase students’ understanding of complex scientific concepts while also overcoming previous scientific misconceptions (NRC, 2005; Schwarz and White, 2005; Tripto et al., 2013). In the sciences, students are asked to make their thinking explicit by constructing or manipulating conceptual, physical, or computer-based models (Vattam et al., 2011; Dauer et al., 2013; Cooper and Oliver-Hoyo, 2017; Howell et al., 2019; King et al., 2019). Computer-based (i.e., computational) models describe relationships between components mathematically, and these relationships can range in complexity from discrete, probabilistic descriptions to continuous, kinetic descriptions. Computational models leverage the power of computers to visualize and study the dynamics of complex systems that are encountered in most scientific disciplines. Because computational models rely on mathematical descriptions, they are well suited to help students adopt a systems-thinking perspective. When used in the classroom, computational models can help students simultaneously hone practical skills, increase content knowledge, and overcome scientific misconceptions (NRC, 2005; Streicher et al., 2005; Riess and Mischo, 2010; Martinez et al., 2011; Liang et al., 2012; Tripto et al., 2013; Bergan-Roller et al., 2018; Hester et al., 2018; King et al., 2019). Computational models of complex biological and biochemical processes and simulations can, therefore, actively engage students in an experiment-like learning environment (Bayraktar, 2001; Riess and Mischo, 2010; Rutten et al., 2012). Moreover, using models to predict experimental outcomes is a recognized learning goal for biochemistry students (AAAS, 2011; NRC, 2013; White et al., 2013).
Although the learning benefits of using computational models in other scientific disciplines have been documented, learning outcomes and effect sizes from computer-based instruction can vary within and across disciplines (Bayraktar, 2001). There may also be differences in how male and female students interact with technology-based instruction, which could affect their learning (Young, 2000; Heemskerk et al., 2009). Computational models are commonly used by biochemists and medical professionals in research and practice. Consequently, it is crucial to understand how computer-based approaches can be used in biochemistry classrooms to introduce relevant technical skills without sacrificing essential content while also maintaining equitable learning. Moreover, despite the documented need to learn about metabolism’s integrated nature, few reports have measured learning gains when biochemistry students receive explicit, interactive practice with integrated metabolic systems.
We hypothesized that teaching metabolism using interactive computer simulations combined with explicit systems-thinking prompts would increase biochemistry students’ mechanistic understanding of complex metabolic systems. We defined mechanistic understanding as the ability to recognize how individual interactions between components affect the behavior of a system under different conditions. The computer simulations were designed with an interactive activity (i.e., module), and we used the predict–observe–explain (POE) instructional model to structure the computer simulation modules (Kearney et al., 2001). By using the POE instructional model, we provided students with the opportunity to practice systems thinking by framing the system as a collection of components that must be carefully understood and analyzed to explain the system’s responses.
To target specific learning objectives that were aligned with ASBMB learning goals, we designed and tested two computer simulation modules in two upper-level biochemistry courses: 1) Regulation of Cellular Respiration (a familiar system; Table 1) was presented in Biochemistry I during the Fall semester, and 2) Regulation of Purine Biosynthesis (an unfamiliar system; Table 2) was presented in Biochemistry II during the Spring semester. During the Fall semester (Biochemistry I), we compared assessment results from the Regulation of Cellular Respiration module to those from a course that received typical classroom instruction (i.e., without the module). During the Spring semester (Biochemistry II), we compared assessment results from the Regulation of Purine Biosynthesis module between students who used modules during the Fall semester and those who did not, and who were all currently in the same course and using the modules. We specifically designed and evaluated modules about familiar and unfamiliar systems and considered students’ previous exposure to the modules to gain additional insight into how the modules impact student learning. We also evaluated the effect of gender when students were using the modules to learn. Together, our results indicate that both modules facilitated students’ mechanistic understanding of complex biological systems, but that repeated exposure may be needed to achieve deep learning when using computer-based approaches to learn about the regulation of an unfamiliar metabolic system, such as the purine biosynthesis pathway.
| Topic and associated assessment | Learning objective | Assessment Item | ASBMB learning goalsa |
|---|---|---|---|
| Glycolysis (assessment 1.1) | 1. Mechanistically explain why and how energy charge affects glycolytic intermediates. | 1a, 1d, 1g, 1h, 1i | 1, 2, 3, 4 |
| 2. Mechanistically explain the role of glucokinase and hexokinase in glucose absorption. | 1e, 1f, | 1, 2, 3, 4 | |
| 3. Contrast mechanisms of regulating glucose absorption to those regulating pyruvate production. | 1b, 1c | 1, 2, 3 | |
| TCA(assessment 1.2) | 4. Mechanistically explain why and how energy charge regulates tricarboxylic acid cycle intermediates. | 2a, 2c, 2e | 1, 2, 3, 4 |
| 5. Mechanistically explain why and how NAD+/NADH redox state regulates tricarboxylic acid cycle intermediates. | 2d, 2f | 1, 2, 3, 4 | |
| 6. Describe the effect of anaplerotic reactions. | 2b, 2g, 2h | 1, 2 | |
| ETC(assessment 1.3) | 7. Mechanistically explain the importance of O2 in cellular respiration. | 3a, 3b, 3j | 1, 2, 3, 4 |
| 8. Mechanistically explain the effect of NAD+/NADH redox state on ATP production. | 3e, 3f,b 3g, 3h | 1, 2, 3, 4 | |
| 9. Mechanistically explain the effect energy charge on ATP production. | 3c, 3d, | 1, 2, 3, 4 | |
| 10. Describe how lactate dehydrogenase maintains glycolysis in the absence of O2. | 3i | 1, 2, 3, 4 |
| Topic and associated assessment | Learning objective | Assessment item | ASBMB learning goalsa |
|---|---|---|---|
| Purine Biosynthesis (assessment 2) | 1. Identify and describe individual interactions that contribute to the regulation of the de novo purine biosynthesis pathway. | 1a,b 1b, 1c, 1d, 1e,b 1f, 1g, 1h, 1i | 3 |
| 2. Mechanistically explain how homeostasis of de novo purine biosynthesis is maintained. | 2a, 2b, 2c | 1, 2, 3, 4 | |
| 3. Describe how changes in cellular conditions affect the metabolic intermediates of de novo purine biosynthesis. | 3a, 3b, 3c, 3d | 1, 2, 3, 4 | |
| 4. Mechanistically explain how mutations in purine biosynthetic enzymes result in metabolic disease. | 4a, 4b, 4c, 4d, 4e, 4f | 1, 2, 3, 4 |
METHODS
Technology
Computer-based learning was facilitated through Cell Collective, a Web-based, research-grade software that makes computational models attainable for use by students and teachers (Helikar et al., 2012, 2015). Students can alter any network or component of the process and instantly observe the effects of the changes made to the modeled system. In the background, computational models in Cell Collective are mathematically described as probabilistic Boolean control networks (Helikar et al., 2008; Abou-Jaoudé et al., 2016). These models consist of components connected with arrows. Each component can represent a variety of elements ranging from a single enzyme or metabolite to an entire process, depending on the scope of the model and the level of abstraction (Helikar et al., 2012). The arrows represent direct or indirect interactions among the components, and students can add or remove components and arrows to observe the effects of changes. To define the regulatory mechanism of a component in Cell Collective, students can add its activators, inhibitors, or a combination of activators and inhibitors via the software’s drag-and-drop user interface. Students can also build a component’s conditional relationships if needed.
The model inputs and outputs are semiquantitative and describe a particular model component’s relative activity in response to environmental signals or perturbations in the model (Helikar et al., 2012, 2015). Users can define the activity level of any external component (input) on a scale from 0 to 100, representing the percent chance of the external component being active or inactive at any time during the simulation (Helikar et al., 2012). The overall activity of any internal component (output) of the model also spans the 0 to 100 scale and represents the average activity (fraction of ones) over a defined number of previous time steps (Helikar et al., 2012). For instance, if a component has an activity level of 50%, it means that the component assumed the same number of active and inactive states over the last n number of iterations, which can be likened to the concept of “moving average” (Helikar et al., 2012).
For an illustration of the practical implications of how changing the model’s connections would change the model’s output, glucose-6-phosphate dehydrogenase (G6PDH) can be used as an example. Glucose 6-phosphate and NADP+ must both be present for G6PDH to be active. These regulatory relationships are qualitatively described as follows in Cell Collective: the G6PDH component is activated by the glucose-6-phosphate component IF/WHEN NADP+ is also active. If students were now asked to test the hypothetical effect of ATP directly inhibiting G6PDH, they would add a negative regulatory relationship between the ATP component and G6PDH component. Consequently, the average activity of the G6PDH component would be reduced over time, because every time the ATP component is active, it inactivates G6PDH.
Access
Each of the modules can be accessed in full via Cell Collective by visiting https://cellcollective.org and selecting “Learning—Get started” on the home page (i.e., no installation is needed). Users can access all content without registration, and users who create free accounts can save their work. The presentation slides we used to introduce the modules and facilitate peer instruction are provided as a supplement to this publication (Supplemental Files S2 and S6). Assessments are also provided (Supplemental Files S4 and S8), and assessment keys are available upon request from the authors.
Module Design
The first module, Regulation of Cellular Respiration, consisted of three sections: Glycolysis (assessment 1.1), the Tricarboxylic Acid Cycle (assessment 1.2, hereafter called TCA), and the Electron Transport Chain and Fermentation (assessment 1.3, hereafter called ETC). The module addressed 1) how the energy charge status and redox state of the cell regulate glycolysis and the tricarboxylic acid cycle, 2) how the electron transport chain and fermentation are integrated into this system, and 3) how the system maintains homeostasis despite changes to the environment (e.g., oxygen availability; Supplemental Files S2–S4). The second module, Regulation of Purine Biosynthesis (assessment 2) addressed 1) how the regulatory mechanisms of the purine biosynthesis pathway allow the cell to maintain homeostasis despite changes to the environment and 2) how mutations disrupt the cell’s ability to maintain homeostasis (Supplemental Files S6–S8).
Instructors and researchers typically emphasize the benefits of having students build, evaluate, and revise their own models as opposed to using expert constructed models (King et al., 2019; Gobert and Buckley, 2000). However, due to the complexity of the systems under study and the time that would be required to fully model and troubleshoot the behavior of the systems, we elected to use an intermediate “model elaboration” approach. For the model elaboration approach, we provided students with key components and asked them to add in known regulatory relationships and then reason through the effects of these individual relationships on the function of the entire system (Bergan-Roller et al., 2018). Using the learning objectives as a guide, we developed computational models up to 3 months before the planned class to allow sufficient time to adjust the module and optimize the models to fit the module design. We used textbook sources to identify critical components to be included in each computational model and manually curated published evidence for regulation. When available, we also used published literature to confirm model outputs. All our models were fully annotated with literature references in the Cell Collective software.
We also created a series of interactive activities that provide students with informational prompts, instructions, and questions as they interact with the software’s model-building and simulation components (Figure 1). For example, in Regulation of Cellular Respiration: Glycolysis, students are provided with a partially built model that is missing important allosteric feedback relationships. Students can edit the model and then predict and evaluate what effect their edits will have. Finally, they can simulate the model’s behavior to test whether their predictions were accurate.
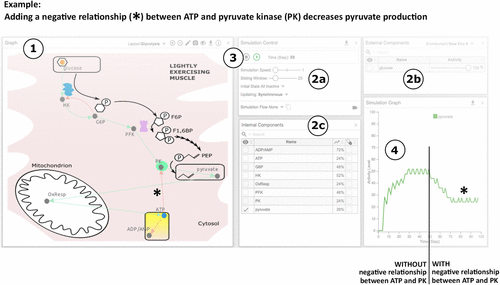
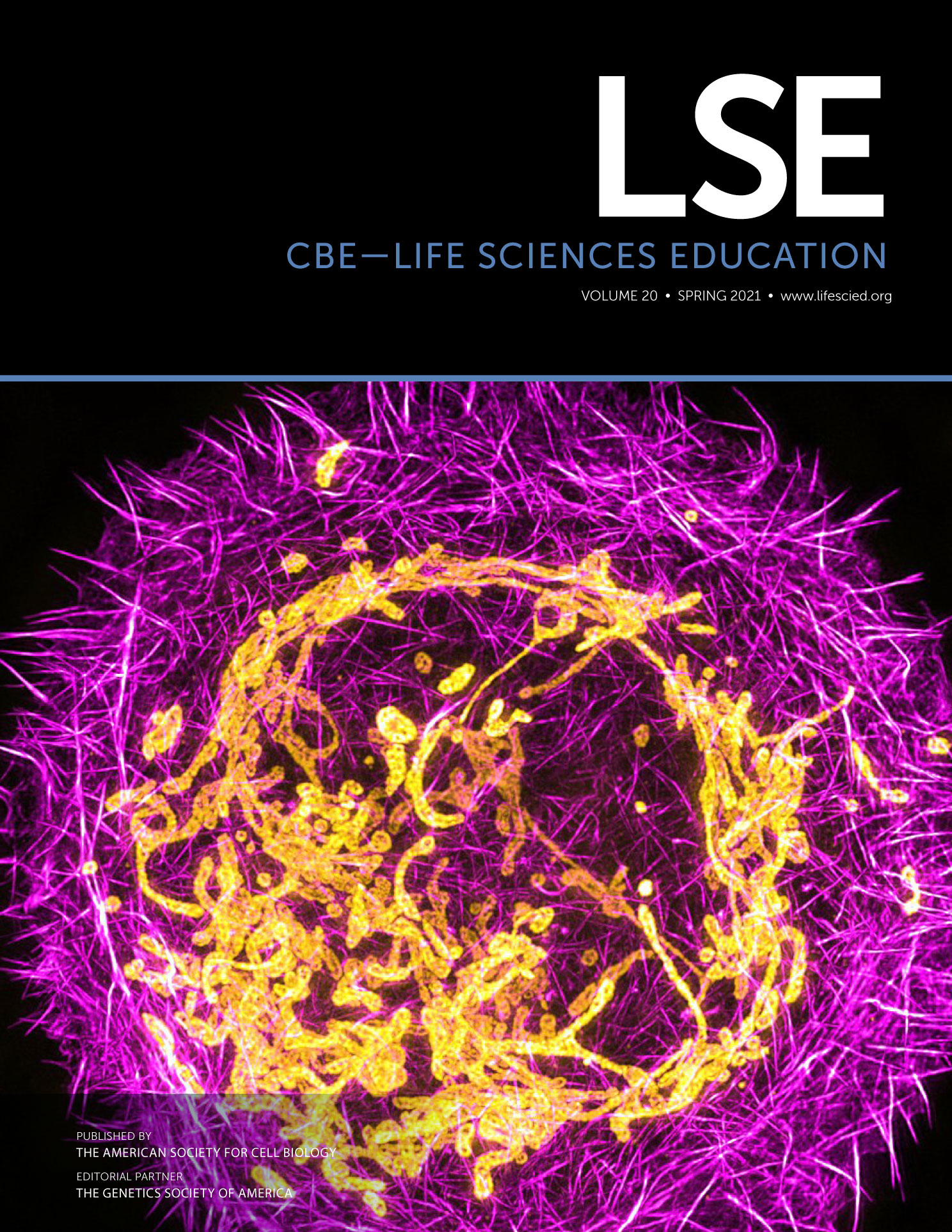
FIGURE 1. Computer simulation modules allow students to adopt a systems-thinking perspective when using computational models to understand the regulation of metabolic pathways. The Cell Collective Web-based software allows students to have an interactive model-based learning experience in which they can (1) edit the computational model by adding components (gray dots) and/or positive or negative relationships (green or red arrows), (2a–2c) set the simulation parameters, (3) simulate the model's behavior, and (4) evaluate the effect of changing the model or simulation parameters on the model's dynamics. For example, to determine the effect of negative allosteric regulation of pyruvate kinase (PK) by ATP, students can add a negative relationship between PK and ATP (*) and observe that pyruvate production decreases. In this example, students could also change the level of glucose by adjusting the slider and selecting additional components to view in the model by checking the box next to the components.
We used an iterative approach to test and refine the module activities and assessment before incorporating them into the classroom. We conducted a think-aloud exercise with one to four senior biochemistry or graduate students focused on usability testing for each module. During the sessions, we noted what participants were saying and doing to ensure that we were achieving the desired interaction with the module. When it was apparent that participants were struggling, we engaged them directly to understand the source of their difficulties. This process helped us develop activities that could be used as stand-alone assignments to reduce instructor burden and increase benefits for distance-learning students.
Assessment Design and Evaluation
We designed our assessments to be functional measures of the concepts that we targeted in each learning objective (Tables 1 and 2). We used multiple true-false (MTF) questions consisting of a question stem that is presented together with a series of statements that students evaluate as true or false (Supporting Files S4 and S8). We selected MTF questions because they can reveal student misconceptions that remain undetected in free-response and multiple-choice question formats (Hubbard et al., 2017; Couch et al., 2018; Brassil and Couch, 2019). Each question in our assessments was unique, and successive assessments were not designed to evaluate longitudinal learning of a concept (e.g., the “energy charge” concept was measured in the Glycolysis, TCA, and ETC assessments, but the questions were unique to each system). Each assessment was developed by two or three biochemistry experts and subjected to multiple rounds of iterative review, including one or two rounds of review by experts who were not involved in developing the original questions. All instructors also approved the final assessments before distributing them to students.
Our study design and timeline allowed us to test a pilot version of the Regulation of Purine Biosynthesis assessment during a previous year. We evaluated the quality of the assessment by calculating the difficulty and discrimination indices of each item from the assessment. We then conducted student interviews and redesigned the assessment based on student feedback. We also used what we learned from the Regulation of Purine Biosynthesis pilot to develop the Regulation of Cellular Respiration questions with input from students who participated in the think-aloud testing for the modules.
For our final analysis, we evaluated the quality of each assessment by calculating the difficulty and discrimination indices of each item in the final assessment (Supplemental Tables S1 and S2). To determine item indices for the pre- and post-assessment scores, we combined student responses from a course in which modules were used with student responses from a course in which modules were not used (the only exception was the Glycolysis assessment during the first year of Biochemistry I, for which we only evaluated student responses from a course in which modules were used). We were willing to tolerate a few items below the 0.2 discrimination threshold for MTF questions (Couch et al., 2015). However, we scrutinized items with negative discrimination on the post-assessment score more closely. If an item had a negative discrimination score in the final assessment, we confirmed that the assessment would still adequately cover the learning objectives if it were removed. We decided to eliminate all items with negative discrimination values from our analysis and future assessments based on these item reviews.
We did not evaluate the values of Cronbach’s alpha for our assessments, because we did not expect them to be above the typical 0.7 cutoff for two reasons: 1) the Regulation of Cellular Respiration assessments were designed to have fewer items per section (i.e., Glycolysis, TCA, ETC) so that the combined pre-assessment was not overwhelming for students, and 2) there is not necessarily a theoretical basis for the items in any of the assessments to comprise a single construct (Adams and Wieman, 2011).
Implementation
We implemented the computer simulation modules in two large-enrollment senior-level undergraduate biochemistry courses (i.e., the “module” courses). The courses comprise a two-part series (here called Biochemistry I and Biochemistry II) that are typically taken in sequence. For Biochemistry I, we also implemented assessments in a comparable Biochemistry I course that was taught by a different instructor who did not use the modules (i.e., the “no module” course).
For each module, we followed the same general format of 1) pre-assessment, 2) instruction and module activities, and 3) post-assessment (Figures 2A and 3A). To prepare for class, the instructor and teaching assistants completed the module. Approximately a week before students started the modules in-class, they individually completed a Cell Collective training module to familiarize themselves with the technology and modeling concepts. Students in the module courses also completed a closed-ended online pre-assessment before the in-class session (Figures 2A and 3A and Supplemental Files S4 and S8). Students in the no module course completed the same pre-assessment online before discussing the topic during class. At the start of the in-class session with the modules, students were introduced to computational modeling and its relationship to the topic through a minilecture (Supplemental Files S2 and S6). During class, students worked in groups of two to four. We used whole-class clicker questions and peer instruction to ensure that students were on target with major concepts and to identify and resolve misunderstandings and technology issues (Crouch and Mazur, 2001). We required students to complete any unfinished activities as homework within a week of covering the topic during class. In the case of Regulation of Cellular Respiration, students only started the Glycolysis part of the module during class and completed all remaining activities as homework (i.e., TCA and ETC were completed entirely as homework assignments over the course of 6 weeks). After completing the modules as homework, students answered the post-assessment questions online to evaluate their learning gains and also completed a short survey about their experiences with the modules (Supplemental Files S4, S5, S8, and S9). Students in the no module course also answered the post-assessment questions online within a week of discussing the topic during class.
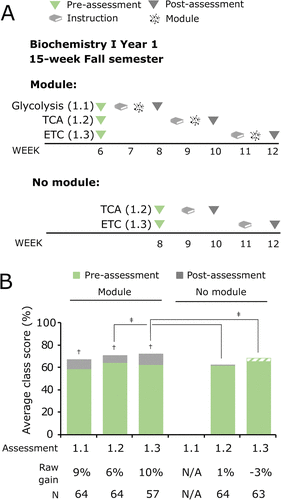
FIGURE 2. Computer simulation modules improve student performance on conceptual assessments of a familiar metabolic system. Assessment and instructional timeline and average class scores for the familiar system of cellular respiration are shown. (A) Diagram of the semester for the module (top) and no module (bottom) courses of Biochemistry I during year 1. (B) Class average values of the pre-assessment scores (green) and post-assessment scores (gray) were compared between module and no module courses for each assessment of cellular respiration (assessment 1.1: Glycolysis; assessment 1.2: TCA; assessment 1.3: ETC). Students in the no module course did not complete the Glycolysis assessment. Each course was taught by a different instructor. Descriptive statistics for raw learning gains are provided in Supplemental Figure S4 and Supplemental Table S5. The average normalized learning gain for each assessment is also provided in Supplemental Table S5. Two-tailed paired t tests were used to measure significance for pre- vs. post-assessment scores: † indicates p < 0.05 (Supplemental Table S5). ANCOVA was used to measure significance for the module vs. no module courses for each assessment: ‡ indicates p < 0.05 (Supplemental Table S6). A green-and-white striped pattern indicates that the overall post-assessment score was lower than the pre-assessment score.
FIGURE 3. Computer simulation modules may improve performance on conceptual assessments of an unfamiliar system if students have previous exposure to similar modules. Assessment and instructional timeline and average class scores for the unfamiliar system of purine biosynthesis are shown. (A) Diagram of the semester for the module course of Biochemistry II during year 1. (B) Students who entered from the Biochemistry I course that used a module during year 1 were designated to be in the second exposure group (46% of students), while students from the no module Biochemistry I course were designated to be in the first exposure group (54% of students). (C) Class average values of the pre- and post-assessment scores for all students in Biochemistry II (“All”) were further subdivided to evaluate the class average values for students in the second exposure and first exposure groups for purine biosynthesis (assessment 2: Purine Biosynthesis). Descriptive statistics for raw learning gains are provided in Supplemental Figure S5 and Supplemental Table S7. The average normalized learning gains for each group are also provided in Supplemental Table S7. Two-tailed paired t tests were used to measure significance for pre- vs. post-assessment scores: ‡ indicates p < 0.05 (Supplemental Table S7). ANCOVA was used to measure significance for the module vs. no module courses for each assessment: ‡ indicates p < 0.05 (Supplemental Table S8).
The instructors of both the module and no module courses introduced the cellular respiration topics in the same sequence (Figures 2A and 3A). However, the module course instructor introduced two lectures about membranes before covering the electron transport chain, while the no module course instructor introduced membranes before beginning glycolysis and completed membranes after covering the electron transport chain. Introducing the modules did not appreciably alter the instructional schedule that was used in previous years.
The Supplemental Material contains additional resources and information necessary to implement the modules, including recommendations for incorporation (Supplemental File S1), slides for minilectures (Supplemental Files S2 and S6), instructor guides (Supplemental Files S3 and S7), assessment questions (Supplemental Files S4 and S8), and student experience survey questions (Supplemental Files S5 and S9).
Data Collection, Participants, and Data Analysis
We collected data in three large-enrollment senior-level undergraduate biochemistry courses at a research-intensive university (Figures 2A and 3A, Biochemistry I module course, Biochemistry I no module course, and Biochemistry II module course). Both Biochemistry I and Biochemistry II are required for biochemistry majors and contain a large pre-health population. Here, we report only the results using data from consenting students for whom we had demographic information and who completed both the pre- and post-assessments.
The Regulation of Cellular Respiration module was implemented in Biochemistry I (N = 107), and the Regulation of Purine Biosynthesis module was implemented in Biochemistry II (N = 142). The module courses were taught by two of the authors (R.L.R. taught Biochemistry I and previously taught the course four times, and K.v.D. taught Biochemistry II and previously taught the material four times). In Biochemistry I, we also compared the learning gains from students in the module course with those of students in Biochemistry I who did not work with the modules (the no module course, N = 121). The no module course was taught by a different instructor at the same university during the same semester as the module course. The instructor of the no module course previously taught the course twice. Biochemistry I instructors from the module and no module courses followed a similar sequence for teaching the course topics and used the same textbook. Both instructors used clicker questions during class, and exams covered the same content and took place during the same week but were independently created and scored. For module courses, each module component (pre-assessment, module activities, and post-assessment) was graded based on completion. For the no module course, the assessments were also graded based on completion.
For the Biochemistry I module course (Regulation of Cellular Respiration), N = 64 for the Glycolysis assessment (assessment 1.1), N = 64 for the TCA assessment (assessment 1.2), and N = 57 for the ETC assessment (assessment 1.3). For the no module course, N = 64 for the TCA assessment (assessment 1.2) and N = 63 for the ETC assessment (assessment 1.3). Logistically, the no module course was unable to complete an equivalent Glycolysis assessment and took the pre-assessment slightly later in the semester compared with the Module course. To ensure reproducibility of the results and to obtain a no module group comparison for the Glycolysis assessment, we repeated the study in Biochemistry I during the following academic year (year 2). During this reproducibility study, the TCA and the ETC assessments were parallel, and both courses were taught by the same instructors as in the first year.
For the Biochemistry II module course (Regulation of Purine Biosynthesis), N = 87 for the Purine Biosynthesis assessment (assessment 2). Approximately equal numbers of students from the module and no module courses from the first year of Biochemistry I were subsequently exposed to modules in Biochemistry II. Students who were exposed to the module in Biochemistry I were designated as “second exposure” students, while students who were not exposed to the module in Biochemistry I were designated as “first exposure” students (Figure 3B). Due to circumstances beyond our control, the second year of Biochemistry II instruction differed dramatically from the first year. To avoid amplification of artifacts, we did not consider those results here.
Participant’s demographic profiles for the module and no module courses in Biochemistry I for both years were calculated in IBM SPSS 23.0 (Supplemental Table S3). We noted a significant difference (p < 0.01) between the average cumulative grade point average (GPA) in the module course (year 1: 3.41, SD = 0.85; year 2: 3.58, SD = 0.35) and no module course (year 1: 3.74, SD = 0.28; year 2: 3.68, SD = 0.29). Similarly, participant demographic profiles for the second exposure and first exposure groups in Biochemistry II revealed a significant difference (p < 0.05) between the average cumulative GPA in the second exposure (3.64, SD = 0.39) and first exposure groups (3.81, SD = 0.18; Supplemental Table S4).
To validate each assessment, we calculated the difficulty index for each item of the assessment in the R programming language (R Core Team, 2013). We then used the discrim function in the psychometric package to calculate the discrimination index for each item (Fletcher, 2015).
To analyze student performance on each assessment, we first calculated each student’s average pre- and post-assessment score for each assessment across all courses or groups (module and no module or second exposure and first exposure). Next, we calculated the mean and SD of the pre- and post-assessment scores for the entire class. Finally, we determined whether students within a course or group significantly improved from pre- to post-assessment by performing two-tailed paired t tests on individual student performance. To calculate the average normalized learning gain, ⟨g⟩, we used the pre- and post-assessment scores for the entire class (Haak et al., 2011). We used a similar approach to analyze pre- to post-assessment score improvements for male and female students and evaluate the significance of student score improvements for individual learning objectives.
We continued our analysis of student performance in the R programming language, where we first calculated each students’ raw learning gain (post-assessment score minus pre-assessment score). Next, we used the ggplot2 package to produce box plots of the learning gains for each assessment or each learning objective (Wickham, 2016). We also calculated the mean, median, and SD of the raw learning gains for each assessment or learning objective for the entire class. We used a similar approach to evaluate male and female learning gains.
Finally, to determine whether there were statistically significant differences between groups, we used IBM SPSS 23.0 to conduct a one-way analysis of covariance (ANCOVA) and performed post hoc pairwise comparisons using a Bonferroni adjustment. Specifically, we compared the post-assessment scores between the module and no module courses in Biochemistry I while controlling for the covariates (pre-assessment score and cumulative GPA) and the demographic variables (gender, native English speaker, parents’ college education, and the extent of education self-funding). We followed the same approach to evaluate the differences between second exposure and first exposure groups in Biochemistry II. We also followed a similar approach when analyzing differences between males and females. However, we first subdivided the student groups based on self-reported gender (i.e., we compared the following four groups: “module and male,” “module and female,” “no module and male,” and “no module and female”). From the ANCOVA, we obtained the values for partial η2, which presents the proportion of variance associated with the main effect after the non-error sources of variation have been partialed out (Richardson, 2011). We used the partial η2 values to estimate the effect size (Richardson, 2011).
RESULTS
Computer Simulation Modules Improve Student Performance on Conceptual Assessments of a Familiar System
To test whether students improved their ability to predict metabolic outcomes after completing the computer simulation modules, we implemented and evaluated the effectiveness of our modules in two upper-level biochemistry lecture courses (Figure 1). The computer simulation modules were designed using the Cell Collective software, and students were asked to predict, observe, and then explain the model’s behavior (Helikar et al., 2012, 2015). Students were prompted to discuss and reflect on their reasoning about the system when they added components or connections to the model.
The first module, Regulation of Cellular Respiration, was integrated into Biochemistry I (15-week Fall semester; Figure 2A). The module was specifically designed to help students understand the regulation of a familiar system, as most students in the course should have learned about this system in one or more previous undergraduate courses. The Regulation of Cellular Respiration module consisted of three sections: Glycolysis, TCA, and ETC (Figure 2A).
To determine whether students achieved learning gains after completing the module, we analyzed class average scores from pre- to post-assessment. Our data show that integration of the Regulation of Cellular Respiration module in Biochemistry I resulted in increased student performance with statistically significant raw learning gains of 9%, 6%, and 10%, for Glycolysis (assessment 1.1), TCA (assessment 1.2), and ETC (assessment 1.3), respectively (Figure 2B and Supplemental Table S5). Conversely, in the no module course, gains were statistically indistinguishable at 1% and −3%, for TCA (assessment 1.2) and ETC (assessment 1.3), respectively (Figure 2B and Supplemental Table S5). To verify the reproducibility of these results, we repeated this experiment in the following academic year (Supplemental Figure S1A). Results for the reproducibility study were consistent with the first year’s results, and students achieved learning gains of 7%, 6%, and 9% in the module course compared with learning gains of −3%, 0%, and 4% in the no module course (Supplemental Figure S1B and Supplemental Table S5). The normalized learning gains for each section of the module were small, indicating a low level of knowledge gain from pre- to post-assessment (⟨g⟩ < 0.3; Supplemental Table S5).
We compared the scores between the module and no module courses while accounting for other factors that could influence these results by using ANCOVAs that included pre-assessment scores and demographic variables as predictor variables (a comparison of student demographic variables for both replicates of Biochemistry I is available in Supplemental Table S3). When we evaluated the post-assessment test scores for Regulation of Cellular Respiration, we found a significant difference between students in the module and no module courses for all sections in both years (year 1: TCA [F(1, 116) = 7.443, p < 0.01, partial η2 = 0.060], and ETC [F(1, 108) = 11.609, p < 0.01, partial η2 = 0.097]; year 2: Glycolysis [F(1, 163) = 6.968, p < 0.01, partial η2 = 0.041], TCA [F(1, 151) = 4.872, p < 0.05, partial η2 = 0.031], and ETC [F(1, 152) = 4.944, p < 0.05, partial η2 = 0.031]; Supplemental Table S6). The partial η2 values we obtained from the ANCOVAs indicate that there was a medium effect for each section of our module for Regulation of Cellular Respiration during year 1, and a small to medium effect during year 2 (partial η2 > 0.06 = medium effect size; Cohen, 1988). These results indicate that the module increases students’ understanding of a familiar topic of metabolism.
Computer Simulation Modules May Improve Performance on Conceptual Assessments of an Unfamiliar System If Students Have Previous Exposure to Similar Modules
The second module, Regulation of Purine Biosynthesis, was integrated into Biochemistry II (15-week Spring semester; Figure 3A). We specifically chose an unfamiliar system that students would not have learned about in a previous undergraduate course. Analysis of student performance on pre- and post-assessments indicated that integration of the Regulation of Purine Biosynthesis module in Biochemistry II increased student performance with a statistically significant raw learning gain of 4% (Figure 3C, Purine Biosynthesis “All,” Supplemental Table S7).
To test whether students’ prior exposure to a similar instructional environment with modules affected their learning about an unfamiliar system, we took advantage of the fact that half of the students in Biochemistry II had experienced the Regulation of Cellular Respiration modules in Biochemistry I the previous semester, while the other half were from the no module group (Figure 3B). We found that students who were exposed to the modules in Biochemistry I (Figure 3C, Purine Biosynthesis, second exposure) achieved a significant raw learning gain of 7%, compared with a non-significant gain of 1% for students who were not exposed to modules in Biochemistry I (Figure 3C, Purine Biosynthesis, first exposure). As in the results for the familiar system, the normalized learning gains for the module were small, indicating a low level of gain in knowledge from pre- to post-assessment (⟨g⟩ < 0.3; Supplemental Table S7).
To compare the scores between the second exposure and first exposure groups, we again accounted for other factors that could influence the results by using ANCOVA with pre-assessment scores and demographic variables as predictor variables (student demographic variables for Biochemistry II are available in Supplemental Table S4). When we evaluated the post-assessment test scores for Regulation of Purine Biosynthesis, we found a significant difference between students in the two groups; F(1, 79) = 8.135, p < 0.01, partial η2 = 0.093 (Supplemental Table S8). The partial η2 value we obtained suggests a medium effect of previously completing a module (partial η2 > 0.06 = medium effect size; Cohen, 1988). These results indicate that students’ prior exposure to computer simulation modules may support subsequent learning with modules, especially when unfamiliar topics or content are being introduced.
Computer Simulation Modules Improve Student Performance on Specific Learning Objectives
To evaluate the modules’ effect regarding specific learning objectives for Regulation of Cellular Respiration in Biochemistry I, we analyzed students’ pre- and post-assessment scores for each objective (Table 1, Figure 4, and Supplemental Table S9). We again confirmed these results during our Biochemistry I reproducibility study (Table 1, Supplemental Figure S2, and Supplemental Table S10). We found that students in the module course generally achieved significant learning gains for at least one objective that represented a focal concept in each section of the module (Glycolysis, TCA, and ETC) and that the learning gain trends were similar between the two years of Biochemistry I (Figure 4 and Supplemental Figure S2).
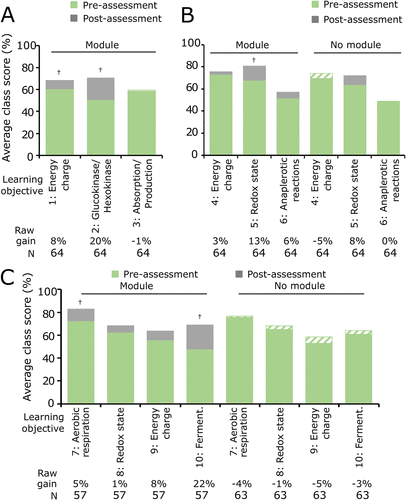
FIGURE 4. Computer simulation modules improve class performance on learning objectives for a familiar metabolic system. Average class scores of the pre-assessment scores (green) and post-assessment scores (gray) for each learning objective for the familiar system of cellular respiration are shown for Biochemistry I during year 1. (A) Assessment 1.1 (Glycolysis), (B) assessment 1.2 (TCA), and (C) assessment 1.3 (ETC) were used to evaluate student learning gains for each objective in the module and no module courses. Each learning objective is numbered, and keywords are provided (detailed objectives are listed in Table 1). Descriptive statistics for the raw learning gains as well as the average normalized learning gains for each objective are provided in Supplemental Table S9. Two-tailed paired t tests were used to measure significance for pre- vs. post-assessment scores: † indicates p < 0.05 (Supplemental Table S9). A green-and-white striped pattern indicates that the overall post-assessment score was lower than the pre-assessment score.
When completing the Glycolysis section of the module, students first investigated the focal concept that energy charge is an important regulatory mechanism of glycolysis (energy charge) and then transitioned into exploring two related focal concepts: 1) the mechanism of how glucokinase and hexokinase affect glucose absorption (“glucokinase/hexokinase”), and 2) how the mechanism for absorption is different from how energy charge–based regulation affects pyruvate production (“absorption/production”). During the first year of the study, students achieved a significant learning gain for the energy charge objective that was covered in the first part of the Glycolysis section of the module. During both years of the study, students achieved mixed results for the concepts that were presented in the second part of the Glycolysis section of the module. The average normalized gains during both years were also small for most of the learning objectives about glycolysis (⟨g⟩ < 0.3; Figure 4A, Supplemental Figure S2A, and Supplemental Tables S9 and S10). We conclude that the Glycolysis section of the module consistently supports small gains in students’ learning about the role of energy charge in regulating the components of glycolysis, the focal concept presented in the first part of the Glycolysis section of the module.
When completing the TCA section of the module, students briefly confirmed how energy charge regulates the components of the tricarboxylic acid cycle (energy charge). However, most of the TCA section of the module focused on two focal concepts: 1) an exploration into the effects of redox-based regulation of tricarboxylic acid components (“redox state”), followed by 2) an investigation into the function of anaplerotic reactions (“anaplerotic reactions”). Although the result was only significant during the first year, we found that the average normalized gain was consistently the largest for the redox state objective, which was covered during the first part of the module (0.7 > ⟨g⟩ > 0.3 for year 1, and ⟨g⟩ < 0.3 for year 2; Figure 4B, Supplemental Figure S2B, and Supplemental Tables S9 and S10). We conclude that the TCA section of the module consistently supports small to medium gains in students’ learning about the role of redox state in regulating the components of the tricarboxylic acid cycle, the focal concept presented in the first part of the TCA section of the module.
When completing the ETC section of the module, students were briefly asked to confirm the integration of energy charge– and redox-based regulation with electron transport chain components (energy charge and redox state). However, the main focus was on concurrently investigating the mechanisms that govern aerobic respiration (“aerobic respiration”) and describing the role of lactate dehydrogenase in fermentation (“fermentation”). During both years, students achieved a significant pre- to post-learning gain for the aerobic respiration and fermentation objectives, indicating that the module consistently supports students’ learning about both focal concepts presented concurrently in the ETC section of the module. The average normalized gains for aerobic respiration and fermentation during both years further indicated that the ETC section of the module consistently supports medium gains in students’ learning about the role of aerobic respiration and fermentation in cellular respiration (0.7 > ⟨g⟩ > 0.3; Figure 4C, Supplemental Figure S2C, and Supplemental Tables S9 and S10).
We did not detect significant gains for learning objectives that were nonfocal concepts. These objectives were typically presented in detail in a previous section of the module and then only briefly reviewed in a later section. For example, students did not achieve significant learning gains for the energy charge objective in the TCA and ETC sections of the module, even though they previously achieved significant gains when this objective was a focal concept in the Glycolysis section of the module. Similarly, students did not achieve significant learning gains for the redox state objective in the ETC section, even though they previously achieved significant gains when this objective was a focal concept in the TCA section of the module. Importantly, in contrast to the module course, no significant differences were detected for any of the learning objectives in the no module course, and all average normalized gains were consistently small (Figure 4, Supplemental Figure S2, and Supplemental Tables S9 and S10).
We also analyzed students’ pre- and post-assessment scores for each learning objective for the Regulation of Purine Biosynthesis module about an unfamiliar system in Biochemistry II (Table 2, Figure 5, and Supplemental Table S11). When completing the module, students began by briefly investigating the individual components and interactions of purine biosynthesis (“components & interactions”). However, most of the module was dedicated to concurrently exploring two related focal concepts: 1) how homeostasis is maintained (“maintaining homeostasis”), and 2) how changes in cellular conditions affect purine biosynthesis (“cellular changes”). Finally, in the last part of the module, students continued to explore cellular changes concurrently with the focal concept of how mutations support disease (“mutations & diseases”). We first evaluated students’ learning gains regardless of previous exposure to modules in Biochemistry I (Table 2 and Figure 5, “All”). Our results show that all students in the module course achieved significant gains for the components & interactions objective, a nonfocal concept introduced at the start of the module. Our results also revealed significant gains for the mutations & diseases objective, a focal concept presented in the last part of the module.
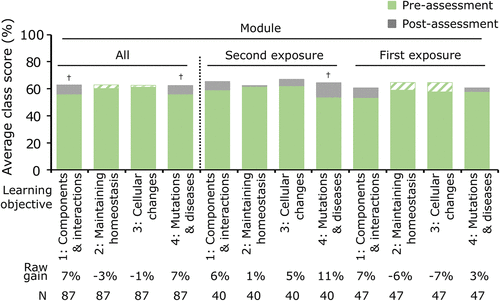
FIGURE 5. Computer simulation modules improve class performance on learning objectives for an unfamiliar metabolic system. Average class scores of the pre-assessment scores (green) and post-assessment scores (gray) for each learning objective for the familiar system of purine biosynthesis are shown for Biochemistry II during year 1. Assessment 2 (Purine Biosynthesis) was used to evaluate all students’ learning gains for each objective (“All” to the left of the dashed line). Students in Biochemistry II were further subdivided to evaluate the class average scores for students in the second exposure and first exposure groups (to the right of the dashed line). Each learning objective is numbered, and keywords are provided (detailed objectives are listed in Table 2). Descriptive statistics for the raw learning gains as well as the average normalized learning gains for each objective are provided in Supplemental Table S11. Two-tailed paired t tests were used to measure significance for pre- vs. post-assessment scores: † indicates p < 0.05 (Supplemental Table S11). A green-and-white striped pattern indicates that the overall post-assessment score was lower than the pre-assessment score.
To gain further insight into students’ learning with the Regulation of Purine Biosynthesis modules, we compared student learning for the second exposure and first exposure groups. Our analysis revealed that student learning gains for the components & interactions objective appeared similar between the two groups, with a raw learning gain of 6% for the second exposure group and 7% for the first exposure group (Figure 5 and Supplemental Table S11). The statistical power of the analysis was reduced by splitting the groups, and the learning gains were therefore no longer significant for this learning objective. However, when we also consider the combined results for this learning objective (Figure 5, “All”), we conclude that students may have benefited similarly in understanding the system’s basic components and interactions. Our analysis further revealed that the learning gains for the mutations & diseases objective appeared different between the two groups, with a raw learning gain of 11% for the second exposure group and 3% for the first exposure group (Figure 5 and Supplemental Table S11). Moreover, the learning gain was only significant for the second exposure group, suggesting that the learning benefit for this objective is only achieved when students have previously completed Biochemistry I modules. In general, the average normalized gains were small when students were learning about this unfamiliar system, similar to what we observed in Biochemistry I (⟨g⟩ < 0.3; Supplemental Table S11). We conclude that the Regulation of Purine Biosynthesis module supports small gains in second exposure students’ learning about how mutations support the development of disease, a focal concept presented in the last part of the module. With the familiar topic in Biochemistry I, we typically observed significant learning gains for the focal concepts presented in the first part of the modules. The observation that this pattern appears to be reversed in Biochemistry II suggests something unique about students’ learning about the mutations and diseases objective that benefits from having previous exposure to the modules. Taken together, our results suggest that the computer simulation modules improved student performance on specific learning objectives.
Repeated Interaction with Computer Simulation Modules May Increase Learning Outcome Equity
Others have shown that technology use in the classroom can increase gender-based differences in technology-based learning outcomes by impacting students’ attitudes, feelings of inclusion, and learning experiences (Young, 2000; Heemskerk et al., 2009). To understand how our modules were impacting male and female students over the course of a semester, we analyzed our results by dividing students in the Regulation of Cellular Respiration module and no module courses by self-reported gender and compared male and female learning gains as the semester progressed. Male students’ average learning gains in the module course trended negatively over the semester, and they achieved a significant pre–post learning gain across the first two module assessments only (Figure 6). Conversely, female students’ average learning gains in the module course trended positively, and they achieved a significant pre–post learning gain for the final assessment in the series only (Figure 6). The trends were repeated in the module course for Biochemistry I during our reproducibility study in year 2 (Supplemental Figure S3). In contrast, neither male nor female students in the no module course achieved significant pre–post learning gains at any point, and both genders appeared to trend in the same direction over the semester (Figure 6 and Supplemental Figure S3). These results suggest that the learning benefit to female students is increased with repeated interaction with the modules over the course of the semester.
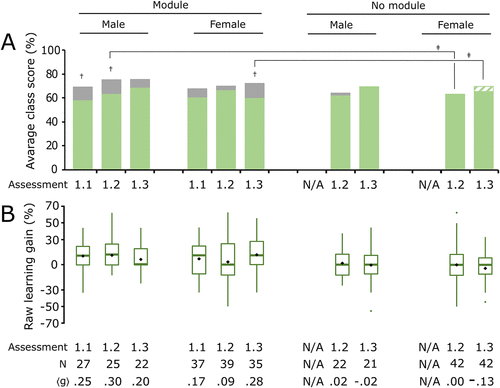
FIGURE 6. Repeated interaction with computer simulation modules may increase learning outcome equity. Average class scores and box plots for individual student learning gains of male and female students for the familiar system of cellular respiration are shown. (A) Class average values of the pre-assessment scores (green) and post-assessment scores (gray) for male and female students were compared between module and no module courses for each assessment in Biochemistry I during year 1 (assessment 1.1: Glycolysis; assessment 1.2: TCA; assessment 1.3: ETC). (B) Box plot showing student learning gains for each group and each assessment. Average normalized learning gain ⟨g⟩ is also shown for each group. Students in the no module course did not complete the Glycolysis assessment. Two-tailed paired t tests were used to measure significance for pre- vs. post-assessment scores of each group for each assessment: † indicates p < 0.05. ANCOVA was used to measure significance comparing the module and male, module and female, no module and male, and no module and female groups for each assessment: ‡ indicates p < 0.05 for a post hoc comparison with Bonferroni correction (Supplemental Table S12). A green-and-white striped pattern indicates that the overall postassessment score was lower than the pre-assessment score. Boxes represent the interquartile range (IQR), and lines within each box represent the median, while diamonds represent the mean. Whiskers represent the highest and lowest values excluding outliers (1.5 times the IQR). Large dark green dots represent outliers.
To confirm that we were not creating or reinforcing a gender bias between male and female groups when implementing the modules, we used ANCOVA to compare the test scores for each assessment between the four groups (module and male, module and female, no module and male, no module and female) after controlling for pre-assessment and other demographic variables. Because the no module course was unable to complete the Glycolysis assessment during the first year, we could not compare those results. For the TCA assessment, we detected a significant difference between the four groups; F(3,115) = 3.021, p < 0.05, partial η2 = 0.073 (Supplemental Table S12). However, post hoc pairwise analysis revealed that only the module and male group was significantly different compared with the no module and female group, indicating that the no module and female group was the only group with significantly lower learning gains. For the ETC assessment, we also detected a significant difference between the groups; F(3,107) = 3.822, p < 0.05, partial η2 = 0.097 (Supplemental Table S12). Specifically, only the module and female group was significantly different compared with the no module and female group, again indicating that the individuals in the no module and female group were the only group with significantly lower learning gains. The results for the second year revealed a similar outcome for the ETC assessment, with female students in the module group trending toward a difference from female students in the no module group. Taken together, the results suggest that the modules did not cause a gender-associated learning gap and could even be improving outcomes for females.
DISCUSSION
Our results support the hypothesis that providing biochemistry students with interactive, guided experiences using computer simulations of metabolic systems (i.e., modules) increases students’ ability to recognize how individual interactions between components affect the behavior of a familiar system under different conditions (i.e., their mechanistic understanding). However, we also found that students’ familiarity with the topic and prior learning with the modules affected their learning.
First, we compared the results for students who learned about a familiar system, Regulation of Cellular Respiration, in a module course for Biochemistry I with results for students who learned about this system in a no module course. We found statistically significant pre- to post-assessment differences, and ANCOVA revealed that incorporation of the module had a medium effect on learning (Figure 2B). We found a similar result when we replicated the Biochemistry I portion of the study, but the effect sizes for the sections of the modules were smaller compared with the first year (Supplemental Figure S1). From these results, we concluded that our modules helped students to develop their mechanistic understanding of a familiar system.
Next, we evaluated the results for students in a module course for Biochemistry II who learned about the unfamiliar system of Regulation of Purine Biosynthesis. Overall, students achieved a statistically significant learning gain from pre- to post-assessment (Figure 3). However, when we further investigated the Biochemistry II results, we found that students who were previously exposed to modules in Biochemistry I (second exposure group) achieved a statistically significant learning gain, while students who were not previously exposed to a module (first exposure group) did not achieve a significant gain (Figure 3). ANCOVA analysis revealed that previous exposure to the modules had a medium effect. We concluded that our modules helped students to develop their mechanistic understanding of an unfamiliar system if they had prior exposure to the modules (Figure 3 and Supplemental Tables S7 and S8). Moreover, we observed lower overall raw and normalized learning gains for the second exposure group in Biochemistry II compared with the module group in Biochemistry I, indicating that students’ familiarity with the system may impact how much learning they achieve with the modules (Figures 2 and 3, Supplemental Figure S1, and Supplemental Tables S5–S8).
Our effect sizes derived from statistical models were consistent with other technology-based learning interventions that have generally been characterized as being effective learning tools (Bayraktar, 2001; Cook et al., 2013). Ideally, effect sizes should be interpreted based on comparable studies rather than typical threshold values, because the effect size will depend largely on the educational context (Bakker et al., 2019; Kraft, 2020). Moreover, outcomes that represent the culmination of years of effort would likely be smaller than typical threshold values (Kraft, 2020). Unfortunately, there is a relative dearth of available effect sizes from the literature for interventions that are used to aid students’ mechanistic learning about complex systems in biochemistry classrooms. As a current best estimate, the comparison of our effect sizes with more general technology-based interventions lends support to the idea that our learning modules were effective (Bayraktar, 2001; Cook et al., 2013). Moreover, because the skills that our modules were targeting should not be expected to develop quickly, our ability to detect medium effect sizes is encouraging.
To understand how the modules facilitated student learning, we investigated student performance on different learning objectives. In Biochemistry I, we found that students in the module course achieved significant learning gains for those learning objectives that directly corresponded to a concept that was the focus of a significant portion of each section of the module about a familiar system (Glycolysis, TCA, and ETC; Table 1 and Figure 4). Furthermore, students tended to achieve learning gains for the same learning objectives during the Biochemistry I replication study. We also found that students’ learning about concepts did not appear to transfer from one section to another when the concept was no longer the focus of a subsequent section of the module. For example, the concept of energy charge was the focus of the first part of the Glycolysis section of the module, but it was only briefly reviewed in the TCA or ETC sections before new concepts were introduced (Figure 4). We found significant learning gains for the energy charge concept for the Glycolysis section of the module (Figure 4A), but not for the TCA or ETC sections (Figure 4B and C). Similarly, students achieved significant learning gains for the concept of redox state in the TCA section of the module (Figure 4B), but not in the ETC section of the module (Figure 4C). Because students’ learning about concepts such as energy charge or redox state did not appear to transfer from one section to another, our results may indicate that students have understood these concepts in the context in which they were presented but have not yet achieved mastery (NRC, 2000). However, students in the no module course did not achieve significant gains in any of the learning objectives, and we concluded that the modules supported students’ learning about important concepts of a familiar system (Table 1, Figure 4, Supplemental Figure S2, and Supplemental Tables S9 and S10).
In Biochemistry II, we first evaluated students’ learning gains for an unfamiliar system regardless of previous exposure to the modules. We found that all students in the course achieved a significant learning gain for the components & interactions learning objective (Figure 5, “All”). This learning objective focused on students’ understanding of the surface features of the system rather than their ability to use principles or synthesis. Conversely, students did not achieve a significant learning gain for the maintaining homeostasis or cellular changes objectives (Figure 5, “All”). Therefore, the result may indicate that students were using a novice approach to analyze the unfamiliar system and had to learn about the specific interactions between individual components rather than understanding how those interactions maintain homeostasis or allow the system to respond to cellular changes (NRC, 2000; Assaraf and Orion, 2005).
We also evaluated Biochemistry II students’ learning gains for different objectives by considering whether students were previously exposed to similar modules in Biochemistry I. The learning gains for the components & interactions learning objective were similar between the second exposure and first exposure groups and the result was significant when both groups were analyzed together (Figure 5, “All”), indicating that the modules provided similar benefits in terms of students’ basic understanding of the system. However, when analyzed separately, we could only detect significant learning gains for students in the second exposure group. Specifically, the second exposure group significantly improved their scores for the mutations & diseases objective after completing the module (Figure 5). To better understand this result, we considered the similarities and differences in the learning history of students in the second exposure and first exposure groups: 1) students in both groups would have previously taken a genetics course, 2) only students in the second exposure group would have previously used the modules and Cell Collective software and would therefore be familiar with the approach, and 3) neither group would be familiar with the purine biosynthesis system. Our results thus support the idea that familiarity with the new learning approach allows students to capitalize on prior knowledge (in this case, information about mutations learned in genetics) compared with students who have not used the learning approach before. One possible explanation is that previous exposure to the modules reduced cognitive load (Mayer and Moreno, 2003). Overall, we concluded that using the computer simulation modules supported students’ learning of important concepts and content of metabolism and that students who are familiar with the modules and software may achieve a better understanding of an unfamiliar system compared with students who use the modules for the first time.
Our results highlight how practical limitations on time could impede students’ ability to achieve mastery. Students will have to master the concept of energy charge to transfer knowledge about it from the Glycolysis section to the TCA or ETC sections in Biochemistry I. They will also need to progress beyond a novice approach to understanding an unfamiliar system before mastering the concept of maintaining homeostasis in Biochemistry II. Because mastery requires a significant investment of time and cognitive resources, we believe it is unrealistic to think that students could achieve mastery after completing a single part of a module. Our modules were explicitly designed with many conceptually challenging questions to ensure that students were engaged in learning for understanding instead of learning for memorization or for improving procedural knowledge (NRC, 2000). However, achieving knowledge transfer of concepts such as energy charge will likely require additional expansion of the modules to provide explicit instruction about applying learned concepts in a different context (NRC, 2000). Additional time would also be required to provide students with explicit opportunities to practice and develop an expert understanding of focal concepts such as maintaining homeostasis in the context of an unfamiliar system. In either case, the additional time investment that would be required was not practical for our courses.
Finally, we investigated potential differences between male and female participants for two reasons: 1) when using technology in the classroom, differences in learning based on gender have been reported (Young, 2000; Heemskerk et al., 2009); and 2) a large study of thousands of biochemistry students showed that females generally underperform compared with males in biochemistry courses (Rauschenberger and Sweeder, 2010). Consistent with Rauschenberger and Sweeder’s observations (2010), we found that female students in the no module course generally underperformed compared with any other group, especially by the end of the semester (Figure 6). Interestingly, our results indicate that repeated exposure to the modules has the potential to make learning gains more similar for male and female students (Figure 6). Further studies are necessary to identify the specific elements responsible for the observed differences in learning gain trends.
Although we cannot formally rule out the impact of the instructor effect with the experimental design that was possible, we provide evidence that supports the effectiveness of the modules: 1) students in the module course and second exposure group had a significantly lower average GPA, but performed better than their counterparts in the no module course and first exposure group; 2) students in the second exposure and first exposure groups in Biochemistry II both had similar pre-assessment scores, but only students in the second exposure group in Biochemistry II achieved significant learning gains overall; and 3) both groups in Biochemistry II were taught by the same instructor. Other study limitations included 1) being unable to collect a comparable data set for a second year of Biochemistry II, and 2) the numbers of male and female students in each subgroup of Biochemistry II being too small to allow us to confirm whether repeated exposure continued to have positive impacts for female students who were previously exposed to the modules in Biochemistry I.
In summary, we designed and tested two computer simulation modules that focus on addressing the conceptual challenges that undergraduate students face when learning about metabolic systems (Brown and Schwartz, 2009). By using approachable computational models that do not rely on the manipulation of mathematical equations, we effectively incorporated computer-based approaches to teaching and learning into an upper-division biochemistry series. Using this approach, we found that early introduction of computer simulation modules improved learning outcomes for undergraduate biochemistry students learning about complex biochemical networks and their regulation. We direct interested instructors to Supplemental File S1 for additional information about students’ perceptions of the learning modules and detailed incorporation recommendations that can accommodate a variety of teaching strategies and course goals. Considering that the use of computational models and simulations is a core component of national education standards in undergraduate life science education (AAAS, 2011), students who use our modules should be better equipped to meet a variety of emergent challenges in the future.
ACKNOWLEDGMENTS
We thank Audrey Crowther, Thuan Long, Jon Dietz, Daisy Guiza Beltran, James Orf, Grant Ozaki, Brady Caverzagie, Ben Plambeck, Olivia Makos, and Dr. Lisa Briona for feedback on the modules. We thank Drs. Edward N. Harris and Xinghui Sun for administering assessments in their courses. This work was supported by the National Science Foundation grant number NSF DUE-1625804 and was classified as exempt from institutional review board review.