The Bacterial Cytoskeleton
INTRODUCTION
One of the pleasures of teaching introductory biology courses is learning new things about old, familiar subjects … such as the differences between eukaryotes and prokaryotes. For a eukaryotic cell biologist, such learning usually entails examining how bacteria function, in ways other than how they replicate and transcribe DNA and how they synthesize protein. I find it interesting, for example, to understand how bacteria maintain their distinctive spherical, rod-like or spiral shapes; or how they make an external cell wall, if they have one; or how they segregate the products of DNA replication faithfully into daughter cells. New answers to these questions are especially interesting because, in my mistaken eukaryote-centric view, bacteria lack cytoskeletons and cytoskeletal proteins, which might be involved in maintaining cell shape, regulating cell wall synthesis, and erecting something like a mitotic apparatus. Which brings me to a second and equally delightful pleasure derived from teaching introductory biology: debunking worn-out notions.
Bacteria do possess cytoskeletons made of proteins which resemble the actin and tubulin familiar to eukaryotic cell biologists. Here I review several, recently published videos that characterize the in vitro behaviors of the actin-like protein, ParM (also known as StbA), and the tubulin-like protein, FtsZ and its in situ localization during cell division. For sake of completeness, I also briefly mention some recent work on the protein crescentin (CreS), an intermediate filament-like molecule, in the absence of published videos.
By way of background material, readers may find the recent review by Michie and Lowe (2006) on the dynamics of bacterial cytoskeletal proteins helpful, including the authors' provocative list of “Future Issues to be Resolved.” Also, students and their teachers may wish to compare the videos reviewed below with those involving tubulin and actin (Watters, 2002, 2004, 2005), which could generate some interesting discussions about the differences and similarities of prokaryotes and eukaryotes. In such discussions, for example, students may raise questions concerning the evolution of “the cytoskeleton”; in which case, they may also find an earlier review on the subject (van den Ent et al., 2001) and commentary (Erickson, 2001) very helpful, for both an overview and a relevant bibliography. Students and their teachers will also want to discuss critically whether the similarities exhibited by the bacterial and eukaryotic cytoskeletal proteins reflect phylogenetic homologies or, rather, represent good examples of convergent evolution.
At this point, readers more familiar with eukaryotic cell biology should be advised the videos being reviewed (as well as the review figures below) were, with one exception, obtained using 100× objectives: that is, at the limits of light microscopy resolution. Thus, the fields of view are small and the images do not seem as large or as sharp as seen in lower-magnification images of eukaryotic cells. Moreover, all but one of the video images were obtained by fluorescence microscopy at low light intensities, which required longer time exposures and time-lapse digital imaging (with accompanying loss of intervening visual detail). In contrast, the set of images portraying FtsZ behavior in vitro was obtained using an atomic force microscope (AFM), which is not a microscope in the usual sense of the term. An AFM lacks lenses and forms an image by means of a probe that traverses the object in a systematic manner, one line at a time. The image formed by the kind of AFM most commonly used for molecular studies is topographical in nature, and image details receive contrast from their size (changes in the z-axis). These images are created in a raster-like manner by the movement of a very fine, whisker-like projection across the surface of an object. AFM resolution, consequently, reflects the size of the probe tip, relative to the detail being probed, and not the diffraction of electromagnetic or electron radiation as seen in more familiar micrographs. With a very small tip, spatial resolution in an AFM image can be very high. Temporal resolution, however, is limited, because of the time usually required to achieve raster-like movements across the field. More information about AFM may be obtained at http://stm2.nrl.navy.mil/how-afm/how-afm.html#General%20concept.
INTRODUCTION TO BACTERIAL CYTOSKELETON
In Escherichia coli, the separation of replicated genomic DNA involves filaments of MreB, an actin homologue (Kruse et al., 2003). In the same species, replicated copies of the R1 drug-resistant plasmid seem to be segregated to opposite poles of a dividing bacterium by filaments of ParM, which is encoded by the plasmid. ParM filaments have been called a “minimalist mitotic spindle” (Garner et al., 2004). Although many students may find this catchy phrase easy to remember, they may also find it misleading, because a ParM filament is not tubular nor does it resemble a prototubular filament. Structurally, ParM, like MreB, resembles G-actin (globular) rather than tubulin, which is the protein subunit of microtubules and the eukaryotic mitotic spindle. The phrase correctly focuses readers' attention on the different roles played by homologous proteins during the course of evolution (and within the same cell, by different paralogues encoded by host cell and plasmid genomes). Thus, in prokaryotes, the separation of replicated DNA is provided by actin homologues (ParM and MreB). Alternatively, as discussed below, division of daughter bacterial cells seems associated with a tubulin homologue (FtsZ) and not an actin-like system (Errington et al., 2003).
Thus, most if not all cells rely on two different groups of cytoskeletal proteins during cell division—one group to separate replicated chromosomes and a second group to effect cytokinesis. Curiously, however, prokaryotes and eukaryotes seem to have taken two different, although complementary, structural routes to achieve the same end.
Thoughtful students will want to know how proteins from organisms so distantly related can be considered homologues, and their skepticism will be heightened when they discover that only ∼15% of the amino acid sequences in actin and ParM (and MreB) are identical. A lively debate might arise about whether proteins containing so few sequences in common can be said to be “homologous.” The debate would be further enhanced by the knowledge that similar “actin folds” are also found in two functionally unrelated types of proteins: sugar kinases (including hexokinase) and the heat-shock protein of 70 kDa (Bork et al., 1992). Unlike actin and ParM (or MreB), however, neither type forms filaments nor, apparently, do they serve a cytoskeletal function. How can such diverse proteins be considered homologous? In this example, homology is based on the unique nature of shared tertiary structure and secondary structural motifs, and how these structures in each protein carry out an identical catalytic activity: the hydrolysis of ATP (Bork et al., 1992). All actin homologues are roughly globular in shape and exhibit a highly characteristic actin fold that consists of two domains hinged at one end and forming a central cleft open at the other end. Both domains in turn contain large and small subdomains with conserved motifs. The fold regulates ATP binding, hydrolysis, and the transient binding of hydrolytic products within the cleft. In ParM and actin, the fold also helps regulate protein assembly and disassembly. The few highly conserved amino acid sequences are strategically located within the nucleotide-binding and hydrolytic creases of the active site.
The filament-forming behavior of ParM, and its nucleotide-binding activities, are of immediate interest, because all known actins are prone to aggregate in a helical and polarized manner (Lodish et al., 2004). Thus, actin filaments (F-actin) consist of two helically entwined subfilaments, which are also polarized. Polarized F-actin has its actin folds oriented in the same direction and parallel with the filament axis, with the cleft of one subunit associated with the hinge of an adjacent subunit (Pollard and Earnshaw, 2004). The filaments are also dynamic, with assembly and disassembly occurring at different rates at the hinge and cleft ends of the filament. The ATP form of G-actin more readily aggregates (and disaggregates) than does the ADP form. Many other proteins associate with F-actin in vivo and regulate its dynamic structure and behavior.
DYNAMIC INSTABILITY OF ParM IN VITRO
The dynamic nature of the ParM polymer, and its relationship to ATP binding by the subunits, has been characterized by Garner et al. (2004), by using total internal reflection fluorescence microscopy and purified ParM labeled with Alexa 488. In the presence of 10 mM ATP, short filaments (Figure 1A) began elongating at both ends symmetrically at approximately equal rates (Figure 1B). Abruptly, polymerization stopped and disassembly occurred unidirectionally and rapidly (Figure 1C). Students with some knowledge of actin polymerization will be puzzled by both observations, because 1) F-actin polymerization proceeds asymmetrically at different rates and 2) ParM depolymerization seemed more catastrophic (and reminiscent of microtubule disassembly) than what is usually observed for F-actin. They will also be puzzled by the video images, which seem to show accretions of label at irregular intervals along the sides of an elongating polymer (especially evident in Figure 1B); these accretions are not discussed by the authors.
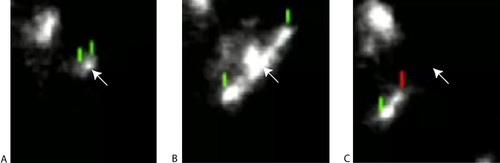
Figure 1. The in vitro behavior of ParM (labeled with fluorescent Alexa 488) incubated in 10 mM ATP. An individual filament begins growing in a bipolar manner (A), and within 10 s, it increases its length more than sixfold (B); in a single frame (5 s), it then depolymerizes catastrophically in one direction to about one-quarter its previous length (C). The green reference marks designate the tips of the growing filament, the red mark designates the shrinking tip, and the arrow represents a fixed reference point. The sequence is taken from a video montage of similar sequences (Movie S4) at http://www.sciencemag.org/content/vol306/issue5698/images/data/1021/DC1/1101313s4.mov. Figure 1 is reproduced from Garner, E. C., Campbell, C. S., and Mullins, R. D. (2004). Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021-1025. Reprinted with permission from AAAS.1
To better characterize the rate of ParM polymerization, the authors also examined filament growth in the presence of adenyl-5′-yl imidodiphosphate (AMP-PNP), a nonhydrolyzable analogue of ATP (Figure 2). Under these conditions, polymerization occurred symmetrically at both ends (Figure 2, B and C) and the polymers grew uniformly longer than in the presence of ATP, and no catastrophic depolymerization occurred. Together, these data suggest that ATP hydrolysis destabilizes ParM filaments, possibly through a conformation change in subunit structure accompanying the release of Pi. The elongation rate constant reported by the authors (∼5 μM−1 s−1) lies between the elongation rate constants reported for the slower and faster growing ends of F-actin (Pollard, 1986).
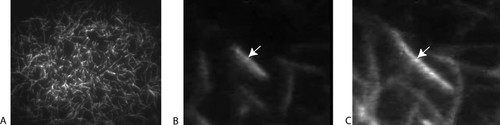
Figure 2. The in vitro behavior of ParM (labeled with fluorescent Alexa 488) incubated in 10 mM AMP-PNP (a nonhydrolyzable analogue of ATP), at low (A) and higher (B and C) magnification. An individual filament grows in a bipolar manner (B) and more than doubles its length in 10 s. The arrow designates a fixed reference point. A was taken from Movie S2 at http://www.sciencemag.org/content/vol306/issue5698/images/data/1021/DC1/1101313s2.mov. B and C were taken from Movie S1 at http://www.sciencemag.org/content/vol306/issue5698/images/data/1021/DC1/1101313s1.mov. Figure 2 is reproduced from Garner, E. C., Campbell, C. S., and Mullins, R. D. (2004). Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021-1025. Reprinted with permission from AAAS.1
More advanced students may wish to examine the rates of ParM nucleation, as determined by fluorescence resonance energy transfer, and the effect of ParM mutations that lacked ATPase activity on polymer stability (Figures 2 and 3 in Garner et al., 2004). These properties can then be compared with similar measurements performed by Garner et al. on F-actin.
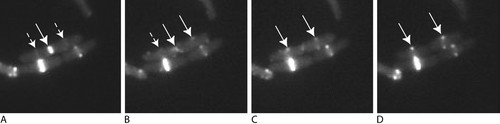
Figure 3. Images showing the division of E. coli expressing GFP-FtsZ, at equal time intervals over a 10-min period. The arrows along the upper, dividing cell designate the so-called Z ring of FtsZ, which forms at the point where cell division occurs: separation into two cells has begun (A), separation is complete (B), and Z rings form in daughter cells in preparation for second round of cell division (C and D). Smaller, broken arrows indicate possible presumptive FtsZ localization. Images were taken from a movie at http://www.cellbio.duke.edu/Faculty/Erickson/pdf's/FtsZmovie.avi. The movie was made by David E. Anderson, Duke University.
Most students who view this and related videos will wonder how ParM (and MreB) separate their respective DNA cargos: directly, through a polymerization/depolymerization mechanism, similar to F-actin in cultured cell locomotion (Pollard and Earnshaw, 2004), or indirectly, by means of motor proteins attached to them and to DNA. Presumably, the direct mechanism effects chromosome separation, but little is known about ParM and MreB ancillary proteins that might provoke another eukaryote-centric response: bacteria lack cytoskeleton-affiliated motor proteins. (Is this next year's outdated notion?)
DYNAMICS OF FtsZ RING ASSEMBLY IN VIVO
FtsZ, an ubiquitous prokaryotic protein, is similar to the eukaryotic cytoskeletal protein tubulin, the subunit of microtubules. As with ParM and actin, the possible homology (descent from a common ancestral gene) of FtsZ and tubulin is based on a similar set of highly conserved features, including 1) tertiary structure, 2) GTPase activity, and 3) ability to form filamentous polymers in vitro (Anderson et al., 2004). FtsZ and tubulin also share a short sequence motif of seven amino acids (Michie and Lowe, 2006). Unlike tubulin protofilaments, however, FtsZ filaments do not form microtubules. Also, as discussed above, ring structures consisting of FtsZ filaments are involved in bacterial cell division and septum formation but not the separation of replicated DNA.
The dynamics of FtsZ ring formation were studied recently by Anderson et al. (2004) by using green fluorescent protein (GFP) chimeras of both wild-type and mutant FtsZ and fluorescence recovery after photobleaching (FRAP) experiments. Although the figures and data of Anderson et al. (2004) were clearly derived from time-lapse digital imaging, none of the sequences was appended to the article. Rather, the Erickson website includes a movie by Anderson that illustrates the behavior of wild-type GFP-FtsZ in a dividing E. coli. For orientation purposes, rings usually form just beneath the plasma membrane in the middle of an elongated cell about to divide. As division occurs, the ring disappears as a complete septum is formed, separating the two daughter cells, and FtsZ disassembles. Later, the two daughter cells each form their own rings as they begin to divide.
The solid bands of fluorescence in Figure 3 represent a condensed band of FtsZ filaments, and Figure 3, A and B, depicts the gradual disappearance of one band in the midline of a dividing cell (central, solid arrow). The ring-like nature of FtsZ filaments was more evident during cell growth (Figure 3, C and D) as two peripheral fluorescent spots that are produced by optically sectioned toruses of labeled filaments. Such rings seemed to begin forming soon after, or possibly before, division of the parent cell (Figure 3, A and B, broken arrows).
When accompanied by appropriate background material, the movie can be understood by introductory biology students, many of whom will note that secondary FtsZ rings did not become prominent until after the primary rings had disappeared. These observations could then be supplemented with a discussion of “precursor pools” and “steady states” and applied to FtsZ. In this context, students might want to predict the effects of inhibitors of protein synthesis on ring formation. More advanced students might want to examine the FRAP data in more detail, to compare the rates of fluorescence recovery in bleached rings with the rates of secondary ring formation after disassembly of the primary ring in the movie. In the absence of a precise time line, any rate estimates from the movie will be crude, although interesting. All students will likely want to learn more about how these tubulin homologues form flexible ring-like structures.
DYNAMICS OF FtsZ RING FORMATION IN VITRO
When studied in vitro, eukaryotic microtubules assemble from tubulin precursors in the presence of GTP (cf. Pollard and Earnshaw, 2004). As GTP is hydrolyzed, some tubules shorten (often catastrophically), whereas others elongate. The tubular structures, however, are stiff and usually straight. Similar studies have not been made on the very transient tubulin protofilaments, but interestingly, electron microscopic images of rapidly depolymerizing microtubules show protofilaments bending as they come apart (Mandelkow et al., 1991).
To study ring formation, FtsZ filaments were created in vitro, by incubating FtsZ briefly in 10 mM GTP, layering small aliquots of the mixture on a mica chip, and washing the adsorbed material with 1 mM GTP (Mingorance et al., 2005). The resulting thin layer was then imaged with AFM as presented in Figure 4. Initially, numerous filaments lay side by side in a linear, mostly straight manner (Figure 4A). With time, the number of filaments decreased and curved bundles formed (Figure 4, B and C). Students should be encouraged to view the entire movie repetitively, tracking changes in filament number and shape in specific regions. It might be useful for them to choose one of the filamentous rings evident in one of the middle frames and then track it backward, to watch how it formed, and forward to observe its disappearance. Of special interest would be an estimate of whether the rings changed their diameter during formation and disappearance.

Figure 4. Dynamic behavior of FtsZ filaments viewed with AFM initially (A), after 10 min (B), and after 26 min (C). Filaments were suspended in a thin film containing 1 mM GTP, and images were captured every 2 min (about the time required to scan the preparation). The same ring is designated by the asterisk; white air-foil-shaped artifacts provide useful fixed reference points. The movie may be viewed at http://www.jbc.org/content/vol0/issue2005/images/data/M503059200/DC1/FtsZPolymersMV.avi.
Although none of the images contains a scale bar, ring diameters could be estimated using the measurements and calculation by Mingorance et al. of an average filament width (∼5 nm). Given this arithmetic, how do these FtsZ rings compare with the ones observed in vivo, if E. coli has a diameter of 800 nm (http://redpoll.pharmacy.ualberta.ca/CCDB/cgi-bin/STAT_NEW.cgi)? Can the range of ring diameters observed in vitro account for the range observed in vivo as an FtsZ ring became constricted and then disappeared? Observant students reading the article will note the filaments were polymerized and washed in a high ionic strength buffer (500 mM KCl, 50 mM Tris, and 5 mM MgCl2), and they might wonder whether the polymerization properties observed under such conditions might vary from those under more physiological conditions, at lower ionic strength (equivalent to 300 mM dissolved ions, including 250 mM KCl and 10 mM MgCl2; http://redpoll.pharmacy.ualberta.ca/CCDB/cgi-bin/STAT_NEW.cgi). Discussion of these conditions could lead to more general considerations of the effects of ionic strength on tubulin polymerization and, more generally, protein structure and function.
CRESCENTIN: AN INTERMEDIATE FILAMENT-LIKE PROTEIN IN CAULOBACTER CRESCENTUS
Some bacteria, notably C. crescentus, exhibit a vibrioid (comma-like) or helical shape, which is thought to be due to the asymmetric localization of filaments formed from CreS, a structural protein recently identified by mutant screening (Ausmees et al., 2003). Filaments composed of CreS labeled with GFP were found exclusively on the inside curves of vibrioid and helical cells, whereas mutant Cres-GFP from rod-shaped cells formed long, sometimes curved filaments some distance from any plasma membrane surface. At the molecular level, CreS exhibits a repetitive, seven-amino acid sequence that is predicted to form coiled-coil structures, and organization of these domains resembles those found in some eukaryotic intermediate filaments such as nuclear lamin A and cytokeratin 19 (see Figure 4 in Ausmees et al., 2003).
I welcome e-mail comments on this article from students and colleagues, especially microbiologists and those who work with cytoskeletal proteins.
ACKNOWLEDGMENTS
Anna Strimaites, a senior Biology major at Middlebury, was immensely helpful searching recent research literature for appropriate videos and preparing the still figures for this article.
FOOTNOTES
1 Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher.



