A Laboratory-intensive Course on RNA Interference and Model Organisms
Abstract
RNA interference (RNAi) is a powerful method to silence gene expression in a variety of organisms and is generating interest not only as a useful tool for research scientists but also as a novel class of therapeutics in clinical trials. Here, we report that undergraduate and graduate students with a basic molecular biology background were able to demonstrate conceptual knowledge and technical skills for using RNAi as a research tool upon completion of an intensive 8-wk RNAi course with a 2-h lecture and 5-h laboratory per week. Students were instructed on design of RNAi experiments in model organisms and perform multiweek laboratory sessions based on journal articles read and discussed in class. Using Nicotiana benthamiana, Caenorhabditis elegans, and mammalian cell culture, students analyzed the extent of silencing using both qualitative assessment of phenotypic variations and quantitative measurements of RNA levels or protein levels. We evaluated the course over two semesters, each with a separate instructor. In both semesters, we show students met expected learning outcomes as demonstrated by successful laboratory experiment results, as well as positive instructor assessments of exams and lab reports. Student self-assessments revealed increased confidence in conceptual knowledge and practical skills. Our data also suggest that the course is adaptable to different instructors with varying expertise.
INTRODUCTION
The importance of a hands-on learning approach has been well established (National Research Council, 2003). The North Carolina State University (NCSU) Biotechnology Education Facility is a core teaching facility designed to offer cutting-edge, hands-on laboratory experiences for undergraduate and graduate students from a range of biological science, chemistry, and engineering disciplines. Students in the Biotechnology Program are required to take a core laboratory course in the manipulation and expression of recombinant DNA (Carson and Robertson, 2006) as a prerequisite for advanced laboratory electives. The RNA interference course described and assessed here is one of the advanced laboratory electives students may opt to take.
RNA interference (RNAi) specifically down-regulates gene expression in a variety of organisms, from tiny single-celled eukaryotic organisms to humans. Although initially discovered in plants (Napoli et al., 1990), work in Caenorhabditis elegans demonstrated that RNAi results from the introduction of double-stranded RNA (dsRNA) into a cell (Fire et al., 1998). The dsRNA interacts with complementary messenger RNA (mRNA) to cause mRNA cleavage and degradation (Paroo et al., 2007; Tolia and Joshua-Tor, 2007). Successful RNAi leads to a decrease in target mRNA levels as well as protein levels (less mRNA means less translation into protein can occur).
RNAi is useful not only as a reverse genetics tool for research scientists but also as a novel approach to a new class of therapeutics; RNAi-based drugs are already in clinical trials (Grinberg, 2008, Lopez-Fraga et al., 2008). In October 2006, Andrew Fire and Craig Mello were awarded the Nobel Prize in Physiology or Medicine for their work in determining the mechanism of RNAi.
Although interest in this field continues to escalate, there is currently no student textbook or laboratory manual in the area of RNAi, and college-level courses are sparse (e.g., Kuldell, 2006). The goal of this course is to combine conceptual knowledge with practical laboratory proficiency to enable students to use this powerful technique. Lectures discuss the history of RNAi and its applications in common model organisms with a focus on the experimental design; journal club papers focus on key historical experiments and are the basis of the laboratory experiments. Multiweek laboratory modules have been shown to enhance student interest and perception of performing experiments in a research setting (Howard and Miskowski, 2005). Here, students perform multiweek RNAi experiments in three laboratory modules, each using a different model organism, to focus on distinct aspects of designing, performing, and evaluating a successful RNAi experiment. In particular, students analyze how the silencing affects an organism on the phenotypic, RNA and/or protein level, which are each important aspects to consider when performing RNAi research experiments.
Learning Outcomes
On completion of this course, students should be able to
design experiments to silence gene expression in various organisms;
generate and understand experiments to assess extent of silencing;
understand the limitations of qualitative and quantitative assessment techniques;
read and present primary literature papers;
use online tools to design RNA silencing constructs to knockdown mammalian protein expression;
describe the advantages and disadvantages of model organisms; and
develop practical proficiency by using Nicotiana benthamiana tobacco plants, C. elegans, and mammalian cell culture.
METHODS
Student Demographics and Participation
Students were not selected to be part of the class but rather were able to freely register upon completion of a prerequisite recombinant DNA lab course within the Biotechnology Program (Carson and Robertson, 2006).
The class section offered spring 2008 consisted of a total of 16 students: five senior undergraduate students (three Chemical Engineering, one Microbiology, and one Zoology), seven M.S. students (four Microbial Biotechnology, one Animal Science, one Forestry, and one Entomology), two Ph.D. students (one Plant Biology and one Entomology), and two continuing education students.
The class section offered fall 2008 consisted of a total of 11 students: three senior undergraduate students (one Chemistry, one Chemical Engineering, and one Microbiology), one M.S. student (Microbial Biotechnology), and seven Ph.D. students (one Chemical Engineering, one Microbiology, one Crop Science, one Entomology, one Plant Pathology, one Functional Genomics, and one Plant Biology).
An anonymous questionnaire addressing the learning outcomes outlined above was given to the students before the first lecture and again after the last lecture, but before the final exam (see Figure 4).
Instructor Information
The class was taught by two instructors. The first instructor (J.A.M.) developed the course and taught the class for five semesters, including the spring course assessed in this study. The second instructor (D.S.W.) assisted in the laboratory for one semester and taught on his own for the fall semester reported in this study. J.A.M. has B.S. degrees in Molecular Biology and in Biochemistry, and a Ph.D. degree in Biochemistry, Molecular Biology, and Cell Biology; D.S.W. has a B.A. degree in Chemistry and a Ph.D. in Molecular and Cellular Pharmacology. Both instructors were teaching postdocs within the NCSU Biotechnology Program who had had previous research experience with mammalian cell culture systems, including RNAi experiments, but minimal to no familiarity in using plants or C. elegans.
Course Synopsis
A laboratory-intensive course on RNA interference was taught over the span of 7.5 wk, followed by a cumulative final exam. Each week consists of a 2-h lecture and a 5-h lab. Laboratory modules used RNAi to knockdown gene expression in N. benthamiana (tobacco plants), C. elegans (soil worm), and mammalian cell culture (human embryonic kidney “HEK”293 cell line). Students silenced a chlorophyll gene in plants and green fluorescent protein (GFP) in C. elegans and in mammalian cell culture cells. The students were responsible for three lab reports, worth a total of 70% of their final grade. The rest of their grade comprised a small interfering RNA (siRNA) design assignment (5%), lecture participation/journal club (5%), laboratory notebooks (5%), and a comprehensive final exam (15%).
The general outline for the lecture schedule was as follows:
Lecture 1: Introduction to RNAi: brief history, endogenous roles, molecular mechanism, applications, and methods for detecting silencing. | |||||
Lecture 2*: Genetics and posttranscriptional gene silencing in plants (Kjemtrup et al., 1998). | |||||
Lecture 3*: C. elegans: model organism and RNAi experiments. (Timmons et al., 2001). | |||||
Lecture 4*: Genetic manipulations and RNAi in Drosophila (Zamore et al., 2000). | |||||
Lecture 5*: RNAi and mammalian systems (Elbashir et al., 2001). | |||||
Lecture 6: MicroRNAs. | |||||
Lecture 7: Therapeutic uses of RNAi: applications, limitations, and clinical trials. | |||||
An asterisk (*) indicates that a journal club discussion was included during the lecture. Paper discussed is shown in parentheses after the lecture topic. | |||||
Students performed silencing exercises in the three laboratory modules. The lab modules and duration are shown below; during three of the weeks, students completed parts of two modules during a single lab period.
Lab 1: Virus-induced gene silencing of magnesium chelatase (su), a chlorophyll biosynthesis enzyme, in N. benthamiana tobacco plants by using microparticle bombardment. Phenotypic and RNA analysis of silencing (5 wk total). | |||||
Lab 2: Knockdown of transgenic GFP expression in C. elegans via feeding of silencing constructs. Qualitative phenotypic assessment of silencing (2 wk total). | |||||
Lab 3: Silencing transgenic enhanced GFP (EGFP) expression in HEK293 cells by using transiently transfected short hairpin RNA expression plasmids. Evaluation of silencing by phenotype and protein expression (3 wk total). | |||||
Laboratory Protocols
Students worked in pairs to perform all laboratory exercises. At the conclusion of a module, students turned in individual lab reports where they applied critical thinking skills to analyze the data in terms of their expectations and how it relates to the data from others in the class and in literature (as appropriate). See Supplemental Material 1 for student lab report guidelines and Supplemental Material 2–4 for instructor lab report grading guidelines.
Introductory Lab
Students purified DNA plasmids, to be used in either Lab Module 1 or 3, with an Endotoxin-free QIAfilter Plasmid Maxi Kit (QIAGEN) and determined their concentration spectrophotometrically.
Lab Module 1: Silencing Chlorophyll in N. benthamianaTobacco Plants
In the first lab, each group transplanted four tobacco seedlings (started from seeds 3 wk in advance) into individual pots containing fertilized soil. Students read and discussed a key paper from the Robertson lab (Kjemtrup et al., 1998) in a journal club format the week before initiating the bombardment experiment. Students used a PDS-1000/He particle delivery system (Bio-Rad Laboratories, Hercules, CA) to initiate virus-induced gene silencing in N. benthamiana to knockdown expression of magnesium chelatase (su), a key enzyme in chlorophyll biosynthesis (Kjemtrup et al., 1998; Peele et al., 2001; Muangsan and Robertson, 2004). Each group bombarded one N. benthamiana plant with a tomato golden mosaic virus (TGMV) genome (5 μg each of TGMV A and TGMV B) as a control and then infected three plants with a virus carrying a 154-base pair su sequence (5 μg each of TGMV A and TGMV B::su) to instigate silencing. The TGMV strain, kindly provided by the Robertson lab, is not infectious. Students monitored the plant height and the number of affected leaves over the following 3 wk. Successful silencing resulted initially in yellow circular spots on the infected leaf and consequently in a spreading of chlorophyll silencing to the new growth of the plant (Figure 1B). Class data were collected 2 and 3 wk after infection and made available to the students.
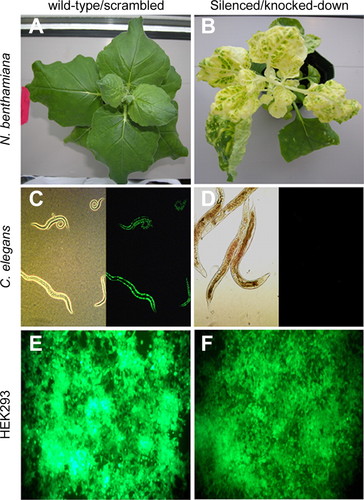
Figure 1. Representative qualitative results from the laboratory experiments. (A and B) Knockdown of su (magnesium chelatase) in N. benthamiana tobacco plants. (C and D) Knockdown of the gfp transgene product in C. elegans. (E and F) Knockdown of EGFP in HEK293 cells constitutively expressing egfp. Knockdown of su (A) or GFP/EGFP (C and E) by using a control (nontargeting) sequence is the panels on the left, whereas specific knockdown of su (B) or GFP/EGFP (D and F) is shown on the right.
Two weeks after infection, each group prepared RNA from 50 to 75 mg from a nonsilenced (green) and a silenced leaf using an RNeasy mini kit and the optional DNase digestion kit (both from QIAGEN, Valencia, CA); A260 readings were used to determine concentration. Students then quantitated su and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels in the silenced and nonsilenced samples by using real-time reverse-transcriptase polymerase chain reaction (RT-PCR) using the iScript One-Step RT-PCR kit (Bio-Rad Laboratories) and the protocol recommended by manufacturer; GAPDH was used to normalize the amounts of RNA present. The primers for the su reactions were Su334-F (5′-GAT CCA AAG ATT GGA GGT GTG ATG-3′) and Su510-R (5′-GCT CCT CAA TTT GTC ACG GAC-3′) and for the GAPDH reactions, GAPDH-F (5′-GAG GTT GCC AAT ATC GTA AGC-3′) and GAPDH-R (5′-CGG TGT AAG AGT GAG TTG TTG-3′). Reactions contained 100 ng of RNA in duplicate or a four-step 1:10 dilution series from the nonsilenced sample to test pipetting accuracy and experimental dynamic range. The following week, the students analyzed the real-time RT-PCR results by applying the comparative Ct method (Applied Biosystems, 2003). Class data were gathered for comparison to individual results.
Lab Module 2: Silencing Transgenic GFP Expression in C. elegans
The week before beginning this module, students read and discussed the Timmons et al. (2001) article upon which these experiments are based. Students investigated which conditions most effectively silence transgenic gfp expressed in C. elegans PD4251 strain, obtained from the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN). C. elegans were maintained on OP50 bacteria on nematode growth media (NGM) for 2–3 wk before the module; see Hope, 1999 for methods to maintain a C. elegans population. In this exercise, students fed the worms two different bacterial strains, BL21(DE3) or HT115(DE3), that either contained or lacked an RNase-specific for dsRNA, respectively. The bacteria had been transformed by the instructor with an empty plasmid (L4440) or one (L4417) with an inducible gfp dsRNA (fresh transformations worked best; data not shown). In the first week, students tested the above-mentioned bacterial strains as well as the best delivery technique: chunking to wet plates or hand-picking to dry plates. Students induced actively growing bacterial strains with 0.4 mM isopropyl β-d-thiogalactoside (IPTG) for 2 h at 37°C before plating them in the center of NGM plates containing IPTG and 100 μg/ml ampicillin “and 12.5 μg/ml tetracycline for HT115(DE3) strains”, thus creating the “wet” plates. Students then transferred an ∼0.5- × 0.5-cm chunk of agar containing PD4251 worms onto the wet plate.
While the bacterial cultures were inducing, the students used picks with platinum wires to individually select and relocate C. elegans to plates that had been seeded with the transformed bacteria by the instructor 4 d previously (“dry plates”). The dry plates were made by the instructors by first pouring NGM plates with IPTG and antibiotics 4–7 d (plates were maintained at room temperature) before seeding with bacterial strains that were induced in the same manner as described above; seeded plates were incubated at room temperature for 4 d before the lab session. Each group hand-picked C. elegans onto a plate of “normal” food source, OP50 bacteria, for practice and to have a nonsilenced control. Students predicted the level of silencing expected for each of the bacterial strains used as a food source. Wet and dry plates incubated at 20°C for 1 wk.
The following week, students observed the C. elegans plates in a fluorescent microscope and visually evaluated the amount of green fluorescence. Students also recorded whether the bacterial lawn was still present because a lack of food will result in a loss of an RNAi phenotype. Students determined which technique (handpicking to dry plates vs. chunking to wet) and condition (bacterial strain) worked best for silencing, i.e., which resulted in greatest loss of green fluorescence, and they compared their results for the different bacterial strains to those seen by Timmons et al., 2001.
Lab Module 3: Knockdown of EGFP Expression in Mammalian Cell Culture
Students read and discussed the first paper describing how to deliver a silencing construct in mammalian cells without eliciting an immune response (Elbashir et al., 2001). The instructors established mammalian cell cultures (HEK293 cytomegalovirus “CMV”-EGFP cells) ∼3 wk before the lab by growing the cells in DMEM-F-12 media supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin/streptomycin; cells were fed approximately every 3–4 d and passaged when near 90–95% confluence (as determined by visual inspection). Using HEK293 cells stably transduced with an egfp gene under the expression of a strong constitutive CMV promoter, students tested the silencing capabilities of three short hairpin RNA (shRNA) constructs cloned into pSilencer 2.1-U6puro plasmid (an siRNA expression vector from Ambion, Austin, TX): two designed (by J.A.M.) to specifically target egfp and the other one was provided from Ambion as a gfp control insert for cloning and an shRNA to wild-type GFP (AmbGFP). The constructs to egfp were siEGFP439, 5′-AACAGCCACAACGTCTATATC-3′ and siEGFP497, 5′-TCAAGATCCGCCACAACATCG-3′ and were chosen using siRNA Target Finder (Ambion), siDesign Center (Dharmacon RNA Technologies, Lafayette, CO), and Whitehead Institute siRNA Prediction Tool (Whitehead Institute for Biomedical Research, Bioinformatics and Research Computing, Nine Cambridge Center, Cambridge, MA). The AmbGFP construct has four mismatches in the area of complementarity with egfp. The day before lab, the instructor plated a six-well plate with 9 × 105 HEK293 CMV-EGFP cells per well. Students delivered 12 μg of the above-mentioned constructs as well as a scrambled control pSilencer construct (Ambion) with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) to the HEK293 CMV-EGFP cells. As controls, one well of the six-well plate received transfection reagent alone and another well neither DNA nor Lipofectamine 2000. Plates were incubated in a humidified chamber at 37°C and 5% CO2. For each of the conditions, students recorded whether they expected no, little, or a lot of silencing. Also during this lab period, students practiced counting cells using extra wells from six-well plates seeded the day before lab by the instructor.
Two days after transfection, students exchanged the transfection media for fresh media in an interim lab period. Plates were returned to the 37°C, 5% CO2 incubator.
One week after transfection, students analyzed silencing by visually evaluating the level of green fluorescence, quantitating the fluorescence using a microplate reader, and performing an immunoblot to assay EGFP protein level knockdown. Students inspected the cells by using a fluorescent microscope and estimated the level of fluorescence compared with the nonsilenced controls. Subsequently, cells were released with TrypLE Express (Invitrogen), counted, and diluted to 2 × 105 cells/ml to be used to quantitate fluorescence and protein levels. To quantitate fluorescence, students plated in triplicate 20,000 and 40,000 cells transfected with pSilencer expression plasmids (scrambled, AmbGFP, siEGFP439, and siEGFP497) in a 96-well plate, and read the fluorescence in a microplate reader. After background correction and averaging like samples, students calculated the percentage of fluorescence remaining by normalizing to the level of fluorescence in the scrambled negative control. Last, the percentage of silencing was calculated by subtracting the percentage of fluorescence from 100. Class data were tabulated and made available for the students; in their lab reports, students examined whether the experiment worked as expected as well as how their microplate results compared with the class data as a whole.
To assess EGFP amounts directly, students prepared protein extract from 6000 cells by using a lysis buffer consisting of 150 mM NaCl, 10 mM Tris, pH 7.4, 1% Triton X-100, 5 mM EDTA, and a Complete mini protease inhibitor tablet (Roche Diagnostics, Indianapolis, IN). Students loaded extract from 1500 cells (alone or with a 1:10 dilution) of the above-mentioned samples along with prestained marker and Cruz Marker MW (Santa Cruz Biotechnology, Santa Cruz, CA) standards on a 10 or 12% polyacrylamide gel. Proteins were transferred to nitrocellulose using a Mini Trans-Blot apparatus (Bio-Rad Laboratories). Blots were stored in Tris-buffered saline (VWR, West Chester, PA) until the next lab period. In the final week, students visualized EGFP through standard immunoblotting techniques by incubating the blot with a 1:3000 dilution of a mouse anti-egfp primary antibody (catalog no. 632381; Clontech, Mountain View, CA) for 60 min and 1:5000 dilution of goat anti-mouse secondary antibody conjugated to alkaline phosphatase (catalog no. sc-2047; Santa Cruz Biotechnology) for 60 min. Students developed the blots by dissolving a nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate tablet (catalog no. 1 697 491; Roche Diagnostics) in water and adding the solution to the membrane. The reaction was terminated with water and blots were photographed.
RESULTS
Laboratory Exercises
Students developed practical proficiency by performing hands-on RNAi experiments using N. benthamiana tobacco plants, C. elegans, and mammalian cell culture. For each module, students were encouraged to critically consider their experimental expectations, class data, and relevant primary literature.
In both semesters, all groups observed knockdown in the expected N. benthamiana plants and C. elegans; sample images of wild-type and silenced plants and worms are shown in Figure 1 (A–D). Students recorded an increase in the number of affected tobacco leaves over time as the silencing spread (Table 1). For the plants, real-time RT-PCR confirmed a decrease in su mRNA expression levels for a silenced leaf compared with a green leaf (Figure 2A); the percentage of knockdown seen by the students varied greatly given that the students could pick a leaf with a large or a small amount of silenced tissue.
| Days postinfection | Construct bombarded | Green | Silenced | Curling |
|---|---|---|---|---|
| 14 | A/B (8) | 68.2 | 0.0 | 31.8 |
| 14 | A/B::su (26) | 50.0 | 43.7 | 6.3 |
| 21 | A/B (8) | 47.0 | 0.0 | 53.0 |
| 21 | A/B::su (26) | 27.3 | 61.0 | 11.7 |
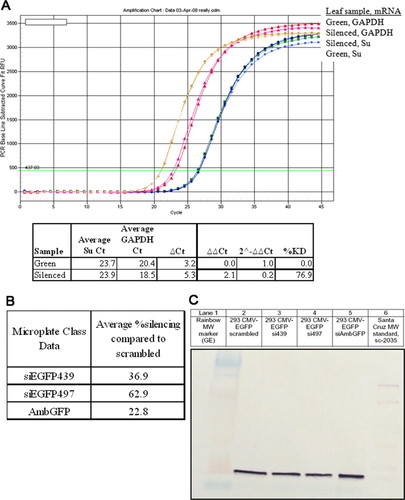
Figure 2. Quantitative results from the laboratory experiments. (A) Sample real-time PCR data and analysis using green leaf sample as a calibrator. The percentage of knockdown varied in class data because leaves showing silencing were variegated (with both green and yellow tissue). (B) Fluorescence intensity from GFP in HEK293 cells measured in a microplate format. Note: GFP is expressed from a very strong promoter (CMV) in the cells. (C) Western blot analysis of protein samples from equal numbers of treated HEK293 cells.
The expression of the EGFP in HEK293 mammalian cell culture cells is very strong due to the CMV promoter, and so a decrease in fluorescence can be difficult to see by eye (Figure 1, E and F); students confirmed the possible phenotype by measuring fluorescence in a microplate reader and by using a semiquantitative Western blot (Figure 2, B and C). In the spring semester, five of eight lab groups observed silencing in HEK293 cells, whereas five of six groups in the fall were able to observe some level of RNAi by measuring the level of GFP fluorescence with a microplate reader. In the spring semester, students loaded protein from 1500 cells and had difficulty determining a clear difference in protein levels; in the fall, students also ran 1:10 dilutions of the samples (150 cells) and were better able to visualize the difference in the EGFP protein levels suggested by the microplate values. Overall, the students observed the most silencing using the siEGPF497 construct and the least with the AmbGFP construct.
siRNA Design Assignment
Students were given an assignment (Supplemental Material 5) to use freely available online tools to choose which of two proposed siRNAs (the effector molecules of RNAi) would be predicted to be more effective in silencing the expression of a particular gene (vimentin; NM_003380). Students were instructed to consider which siRNA better matches published recommendations (e.g., Reynolds et al., 2004) and program parameters as well as which was selected by more than one program. Of the 16 students in the spring semester, 15 correctly identified the better siRNA candidate. Similarly, 10 of the 11 students in the fall semester were able to select the preferred siRNA sequence.
Student Exam Assessment
The final exam consisted of 25 multiple-choice, 13 true/false, and four short answer/discussion questions. The same exam was administered in the two semesters. Both instructors collaborated on grading the spring semester exams and in establishing a rubric to maximize consistency in grading. Figure 3 shows the percentage of accurate responses from students in the spring and fall semesters to the multiple-choice and true/false questions as grouped by lecture topic. The four discussion questions (worth 24% of the grade on the final exam) addressed encompassing topics that were discussed in multiple lectures and incorporated topics from the laboratory exercises; the questions were designed to test critical thinking skills and to allow the students to synthesize their conceptual understanding of RNAi and experimental design. In particular, the discussion questions focused on the learning outcomes dealing with the advantages of model organisms and on designing experiments to silence gene expression in various organisms, including construct selection, controls, and experimental predictions. For the discussion questions, the class average in the spring was 85.0%, with the four individual averages of 68.8, 87.2, 87.2, and 96.6%. For the fall semester, the class average for the discussion questions was 70.5%, with the four individual averages of 60.0, 78.8, 66.7, and 86.3%. The discussion question with the lowest response was a difficult question which required the students to fully integrate the experimental design implications for worms, flies, and mammalian systems. The overall class average for the final was 83.0 ± 7.5% for spring semester and 80.0 ± 10.8 for the fall. Exam questions are available upon request.
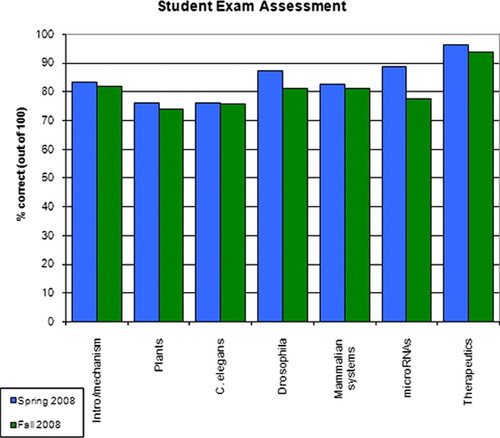
Figure 3. Student performance on final exam. The final exam consisted of 25 multiple-choice, 13 true/false, and four discussion questions. The multiple-choice and true/false questions were divided into categories based on when the topic was primarily addressed in lecture; five to six questions were based on each lecture.
Other Assessment Methods
The instructors evaluated student participation in journal club discussions during lectures 2–5 and student comprehension of silencing experiments in lab reports. Discussions lasted 45–60 min and involved communicating how the experiments were performed as well as interpreting the data. Student participation was judged (on a scale 0–3) to reflect the amount of effort expended in contributing to the discussion, and in meeting the learning objective of reading and presenting primary literature papers. Student lab reports were evaluated for understanding of basic principles of experimental design and theory, as well as interpretation of their data. The lab reports provided a method for assessing the learning goals focused on a student’s understanding of using experiments to assess extent of silencing as well as the limitations of qualitative and quantitative assessment techniques (Supplemental Material 2–4).
Quantitative Student Self-Assessment
Before the first lecture, and again before the final exam, students were asked to anonymously answer a questionnaire to assess their perceived proficiency in both conceptual understanding and technical skills. They were instructed to rate their current level of knowledge or competence for each concept or technical skill on a scale of 1–5, with 1, no knowledge or competence; 2, little knowledge or competence; 3, moderate knowledge or competence; 4, a good deal of knowledge or competence; and 5, excellent knowledge and/or competence. Figure 4 presents the questions asked as well as the responses from pre- and postmatriculation in the class. For both spring and fall semesters, students reported an increase in their level of knowledge or competence in every category tested.
Course Evaluations
Students were encouraged to complete university-wide course evaluations that asked students to respond with 1–5, where 1 was strong disagreement and 5 was strong agreement. Table 2 shows the combined student responses from spring and fall semesters on questions relevant to the course and laboratory. The results indicate that students convey a high degree of satisfaction with the course. Five students in total elected not to participate in the survey.
| Course evaluation | Mean | 5 | 4 | 3 | 2 | 1 | BL | N | Deptmean |
|---|---|---|---|---|---|---|---|---|---|
| This course was intellectually challenging and stimulating | 4.77 | 17 | 5 | 0 | 0 | 0 | 0 | 22 | 4.37 |
| This course improved my knowledge of the subject | 4.91 | 20 | 2 | 0 | 0 | 0 | 0 | 22 | 4.56 |
| The course readings were valuable aids to learning | 4.76 | 16 | 5 | 0 | 0 | 0 | 1 | 21 | 4.32 |
| Lab sessions contributed to mastery of course concepts | 4.72 | 13 | 5 | 0 | 0 | 0 | 4 | 18 | |
| Overall, the labs were effective learning experiences | 4.61 | 11 | 7 | 0 | 0 | 0 | 4 | 18 | |
| Overall, this course was excellent | 4.68 | 15 | 7 | 0 | 0 | 0 | 0 | 22 | 4.39 |
DISCUSSION
Positive Course Outcomes
The goals of this class were to provide students with a conceptual understanding of and practical proficiency in RNAi technology. Lectures and journal article discussions were used to discuss the mechanism and methodologies of RNAi in four model systems: plants, C. elegans, Drosophila, and mammals. In multiweek lab modules, students performed gene silencing experiments in each of these systems with the exception of Drosophila. We assessed the learning outcomes through success in lab experiments, performance on lab reports, an siRNA design assignment, journal club discussion of primary literature papers, student self-assessment questionnaires, the final exam, and class evaluations.
By combining both lecture and literature discussion with the laboratory exercises, the students engaged in active learning and were exposed to the material both in concept and in practice. Student participation in the discussion of primary literature was facilitated by the instructor who projected the figures and asked for student volunteers to describe to the class the type of experiment illustrated or to explain the results or conclusions. Often the procedures and results from the journal articles foreshadowed the experiments that the students would perform in the laboratory. In both semesters, students were able to successfully perform silencing experiments (Figures 1 and 2 and Table 1) and evaluate them in lab reports for the three model organisms, indicating a practical and conceptual proficiency in techniques relevant to each system. Instructor assessment of the final exam (Figure 3) supports that the students gained an understanding of RNAi in the model organisms as well as how it pertains to microRNAs and therapeutics.
The student self-assessments indicated an increase in confidence in both conceptual understanding and in technical skills for both semesters (Figure 4). In general, the largest increases in perceived knowledge/skill were in RNAi experimental design and techniques (questions 5, 8, 12, and 17). The smallest increases were in describing RNAi applications, in reading and discussing primary literature papers, and in manipulating real-time RT-PCR (questions 2, 7, and 14). This is not surprising given that those were knowledge/skill questions to which the students had reported higher perceived proficiency before they took the class. Moreover, 19 of the 27 students are graduate (or postgraduate) students and have had prior experience with reading and presenting primary literature. Overall, the students responded positively to the course readings and to the course as a whole in the course evaluations (Table 2).
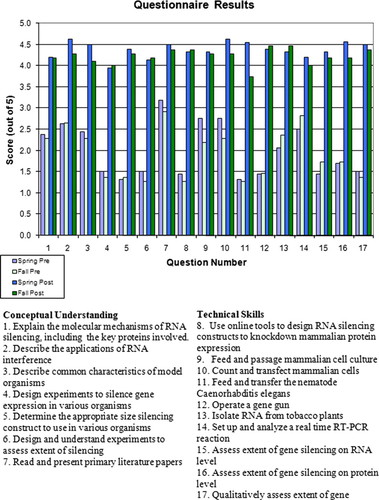
Figure 4. Pre- and postcourse student self-assessment. Data from the quantitative assessment questionnaire was scored and averaged. The asterisk (*) indicates that the “post” data for question 11 (fall) and question 13 (spring) include a score from a student who missed this lab session.
In brief, this course taught by different instructors successfully met its learning objectives. Furthermore, after the completion of the course, one student successfully designed and executed an RNAi experiment in his own research on the American dog tick, Dermacentor variabilis (Mitchell et al., 2007), and has recently accepted a position in a biotechnology company to continue performing research using RNAi.
Course Adaptability
Our data indicate that the course can be effectively taught by different instructors with varying expertise. Neither instructor had had prior experience working with plants or C. elegans; however, a background in molecular biology provided the instructors a foundation to perform RNAi experiments in these systems. The laboratory modules have now been optimized over the course of six semesters and 77 students.
We are confident that the course is adaptable to other institutions. To facilitate other institutions in establishing similar courses, J.A.M. is in the process of writing a textbook, RNA Interference and Model Organisms: in Theory and in Practice, with the RNAi lecture materials and laboratory protocols.
The laboratory component of this RNAi course is subdivided into three modules, any of which could be incorporated individually into existing classes depending on time and equipment available. The modules range from two to five weeks, and here the students performed experiments from multiple modules during three of those weeks. Although the plant RNAi module uses a microparticle delivery system (or gene gun that costs ∼$25,000 to purchase), the protocol can be modified to deliver the silencing constructs using carborundum powder to cause microabrasions on the plants (Ascencio-Ibañez and Settlage, 2007). In the experiments described here, a fluorescent microscope is needed to phenotypically evaluate the silencing of GFP in C. elegans and mammalian cell culture. One could elect to knockdown a different gene, such as unc-22 in C. elegans, which causes uncoordination (Timmons et al., 2001), or focus on the silencing effect on the RNA or protein levels by using real-time RT-PCR or Western blot. J.A.M. integrated an expanded C. elegans silencing experiment into a Molecular Genetics course at Drew University; the students qualitatively assessed the fluorescent effects of knocking down gfp expression and used RT-PCR as a semiquantitative measurement of RNA levels (rather than with real-time RT-PCR). Thus, all three modules need not be taught together; instructors have the flexibility to mix and match components of the modules to fit time and resources available.
In sum, student and instructor assessment of this laboratory-intensive RNAi course suggest a worthwhile learning experience that should be transferrable to other instructors and institutes.
FOOTNOTES
Conflict of Interest: J.A.M. was involved in producing the curriculum and is currently under contract by Jones and Bartlett Publishers to write a textbook covering the material. No promotion of a particular product to the exclusion of other similar products should be construed. J.A.M. and D.S.W. were the instructors of this course.
ACKNOWLEDGMENTS
We acknowledge Dr. Niki Robertson and her laboratory (NCSU), the Caenorhabditis Genetics Center (University of Minnesota), Tranzyme Pharma (Durham, NC), and the Morimoto laboratory (Northwestern University, Evanston, IL) for organisms and reagents. We thank our lab manager, Melissa Cox, for time in ordering and preparing reagents for the laboratories. We also thank the students for effort and willingness to participate in this study. Finally, we thank the NCSU Biotechnology Program for support of this course.



