Can Genetics and Genomics Nursing Competencies Be Successfully Taught in a Prenursing Microbiology Course?
Abstract
In recognition of the entry into the era of personalized medicine, a new set of genetics and genomics competencies for nurses was introduced in 2006. Since then, there have been a number of reports about the critical importance of these competencies for nursing practices and about the challenges of addressing these competencies in the preservice (basic science) nursing curriculum. At least one suggestion has been made to infuse genetics and genomics throughout the basic science curriculum for prenursing students. Based on this call and a review of the competencies, this study sought to assess the impact of incorporation of genetics and genomics content into a prenursing microbiology course. Broadly, two areas that address the competencies were incorporated into the course: 1) the biological basis and implications of genetic diversity and 2) the technological aspects of assessing genetic diversity in bacteria and viruses. These areas address how genetics and genomics contribute to healthcare, including diagnostics and selection of treatment. Analysis of learning gains suggests that genetics and genomics content can be learned as effectively as microbiology content in this setting. Future studies are needed to explore the most effective ways to introduce genetics and genomics technology into the prenursing curriculum.
INTRODUCTION
A new set of genetics and genomics competencies for nurses was introduced in 2006 (Consensus Panel on Genetic/Genomic Nursing Competencies, 2006). These competencies were developed in recognition of the fact that we are entering an era of personalized medicine that includes genetic risk assessment for complex diseases (those with contributions from multiple genes and the environment) and tailored therapies based on underlying genetics. The competencies are clearly focused on human genetics (e.g., “Constructs a pedigree from collected family history information using standardized symbols and terminology”; Consensus Panel on Genetic/Genomic Nursing Competencies, 2006) and on bridging the current gap between genomics research and clinical practice (Guttmacher et al., 2007).
Since the publication of these competencies, a number of articles have addressed the challenges and suggested possible mechanisms to bring practicing nurses up to speed in genetics and genomics, as well as ways to incorporate genetics and genomics into the nursing and prenursing basic science curricula. For example, Guttmacher et al. (2007) stress that the basic content of genetics and genomics needs to be contextualized and linked to case studies that involve real patients. They also stress that genetics and genomics should be incorporated throughout the preservice curriculum for nursing students. Similarly, Prows et al. (2005) note that while adding a genetics course to the prenursing basic science requirements may be an obvious solution, it is not practical. Most nursing programs are already packed with requirements and courses, and one isolated course is not likely to fully prepare students for practicing in a genomic era. On the basis of these considerations, they also suggest consistently integrating genetics into courses in the nursing curriculum (Prows et al., 2005). Similarly, Lewis et al. (2006) argue for the integration of genetics content into existing nursing courses. In describing opportunities and examples for incorporating genetics into the nursing curriculum, Lewis et al. (2006) discuss drug selection and dosage for cancer therapy, using molecular markers of the cancer to inform drug decisions. This approach (genetics informing drug decisions) also applies to the selection of antibiotics and antivirals. In many cases, the pathogen is genotyped to assess pre-existing resistance (e.g., rapid polymerase chain reaction [PCR] for the mecA gene of Staphylococcus aureus or human immunodeficiency virus [HIV] genotyping to inform selection of antiretrovirals in HIV therapy; World Health Organization, 2001; Makgothlo et al., 2009; Division of Medical Assistance, North Carolina Department of Health and Human Services, 2010). Thus the general principles of personalized medicine, as driven by genetics and genomics, apply to infectious disease as well as inherited and sporadic genetic diseases of humans.
In a related approach, Kelly (2008) focuses on colon cancer and describes the role of genetics and genomics in informing patient care in familial and sporadic cases of colon cancer. One theme in this discussion is the need for nurses (and all healthcare professionals) to be conversant using the terms of genetics and genomics—namely, gene, allele, genotype, phenotype, and mutation. It is noteworthy that these terms also apply to the diagnosis and treatment of many infectious diseases. For example, methicillin-resistant S. aureus (MRSA) has an antibiotic-resistant phenotype due to specific alleles of specific genes (an underlying genotype), and closer investigation of the genotype and phenotype can inform antibiotic selection for treatment (Shopsin and Kreiswirth, 2001; Makgothlo et al., 2009). In HIV, the virus is often genotyped to assess pre-existing antiretroviral resistance alleles, and the host genotype at the CCR5 allele strongly influences disease progression. It is therefore clear that healthcare professionals need to have a solid understanding of these basic genetics and genomics terms in order to treat patients effectively.
Students in the prenursing curriculum at New Mexico State University (NMSU) take many of their basic science prerequisite courses in the biology department. These include one semester each of introductory biology and lab, public health microbiology and lab, human physiology, and human anatomy and lab. This curriculum does not include a genetics course. While their introductory biology course covers some aspects of molecular genetics (DNA structure, DNA replication, and gene expression), it does not explicitly cover Mendelian genetics or concepts of inheritance and allelic variation. In an effort to respond to the new mandate, genetics and genomics content and its applications were integrated into a prenursing microbiology lecture course at NMSU. Student mastery of genetics content was assessed by measuring normalized learning gain (using a pre- and post-genetics content test; Hake, 1998). Student attitude toward specific aspects of the course was also assessed. Based on the genetics and genomics implementation and analysis, it appears that prenursing students can learn relevant genetics and genomics content within the context of an introductory allied-health microbiology course.
MATERIALS AND METHODS
Biol 219, Public Health Microbiology
This is a three-credit lecture course designed for prenursing and allied-health majors (e.g., medical technologists, physical therapists, medical assistants, and dental hygienists). The typical enrollment is between 50 and 80 students, the vast majority of whom are prenursing students (see Table 1 for demography of the three semesters of this study). Students enroll in a separate two-credit lab that is shared between the allied-health students and the general microbiology course taken by biology and microbiology majors. The lab and lecture courses are taught independently, by independent instructors, for separate grades. This study focuses on lecture-based instruction and assignments, as not all students take the lab concurrently with lecture, and the lab and lecture courses are not intentionally integrated with respect to particular content areas. Successful completion of one semester of introductory biology is a prerequisite for Biol 219 (Public Health Microbiology). The content of the Biol 219 lecture is consistent with American Society for Microbiology (ASM) recommendations for an allied-health course (ASM Curriculum Recommendations, 2001).
| Gender | Race and ethnicity | Class standing | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Semester | Female (n) | Male (n) | White (n) | Asian (n) | Hispanic (n) | Other (n) | Black (n) | Freshman (n) | Soph. (n) | Jr. (n) | Sr. (n) |
| Fall 2007 | 46 | 4 | 14 | 0 | 24 | 4 | 1 | 2 | 19 | 19 | 10 |
| Spring 2008 | 37 | 10 | 16 | 0 | 24 | 4 | 1 | 0 | 27 | 12 | 7 |
| Fall 2008 | 51 | 8 | 24 | 0 | 24 | 4 | 2 | 2 | 24 | 24 | 8 |
Genetics and Genomics Instruction
Genetics and genomics topics were introduced to the course over three semesters (Fall 2007, Spring 2008, and Fall 2008). In each semester, one of four (Fall 2008) or five (Fall 2007 and Spring 2008) units of the course was designed to teach aspects of genetics and genomics. In each semester, the unit of instruction included interactive (clicker-enhanced) lectures, in-class activities and discussions, and a culminating take-home assignment. The general topics addressed in the genetics and genomics unit included the biological basis of genetic diversity (e.g., genotype, phenotype, alleles, mutations, horizontal transfer in bacteria, and genetic drift and shift in viruses), implications of genetic diversity in the context of natural selection, and technological aspects of assessing genetic diversity and making genetic-based identifications (e.g., PCR genotyping, pulsed-field gel electrophoresis genotyping, and sequencing) (see excerpts from the Fall 2008 syllabus in the Supplemental Material). While the specific examples and assignments used in the instruction were modified each semester (in an effort to improve teaching and learning), the same concepts were included in each of the three semesters, and the overall genetics and genomics objectives were retained across all three semesters. This report focuses primarily (but not exclusively) on the third semester (Fall 2008), as it represents the best-informed semester in terms of instructional approach, based on experiences in the first two semesters. In the third semester, the take-home assignment focused on HIV infection. This permitted explicit incorporation of human genotypes (CCR5 genotyping based on PCR analysis) as well as pathogen genotypes (and sequencing to establish HIV genotypes) and implications for treatment and resistance (concepts of genetically diverse populations and natural selection). See the Supplemental Material for the associated genetics unit take-home assignment for the Fall 2008 semester.
Content Knowledge Assessment
The research protocol and associated instruments were approved by the NMSU Institutional Review Board following an expedited review process (Protocol Numbers 6511 and 326). To assess student learning of genetics and genomics material, students were invited to take a pretest within the first week of the course. The pretest included a variety of multiple-choice, short-answer, and matching questions addressing the biological basis and implications of genetic diversity and genetics and genomics technology (e.g., assaying genetic diversity and genetic-based identification). In the last two semesters, it also included a set of questions addressing more general microbiology topics, in order to provide a comparison topic. These same questions were then embedded on the corresponding midterm exam(s) or final exam, allowing the calculation of a normalized learning gain (Hake, 1998) for the three major themes: biological basis and implications of diversity (Bio), genetics and genomics technology (Tech), and general microbiology (Micro; see the Supplemental Material for the Fall 2008 pretest, representative scoring rubrics, and pre- and posttest results for representative Bio and Tech questions). A normalized learning gain was calculated for each student ([Posttest score − Pretest score] / [Maximum possible − Pretest score]; Hake 1998); then a class average gain (<g>) was calculated by averaging the individual student gains. The normalized learning gain permits normalizing for different pretest scores by calculating the actual gain as a proportion of the total possible gain, based on the pretest score and the maximum possible score (Hake, 1998).
A variety of statistical methods were used to analyze the learning data to test for 1) differences between pre- and posttest scores, 2) differences between topics (Bio, Tech, and Micro) within a semester, and 3) differences within topics across semesters. To look at differences between pre- and posttest scores for each semester, a one-tailed paired sample t test was used to determine whether the student posttest scores were significantly different (higher) from the student pretest scores.
For the Fall 2008 semester, a one-way repeated measures analysis of variance (ANOVA) was used to test for differences in the learning gains among the three content areas (Micro, Bio, and Tech). The content-area gains were calculated by characterizing each question (or question part) as pertaining to each topic and then calculating a pre- and posttest score for each topic. From this, normalized learning gains for each topic were calculated for each student and used in the ANOVA. This overall analysis was then partitioned with contrasts to test for differences in learning gains between 1) Micro and Bio, 2) Bio and Tech, and 3) Micro and the average of Tech and Bio.
To compare the learning gains for each genetics and genomics subtopic (Bio and Tech) across the three semesters (Fall 2007, Spring 2008, and Fall 2008), one-way ANOVAs were carried out for each topic over all three semesters.
Attitudinal Assessment
Student attitude toward and confidence in genetics and genomics was assessed using a two-part end-of-semester anonymous student evaluation. The first part consisted of a focused open-ended question on the end-of-semester student evaluations. This question asked students for their comments on how well the course helped them meet the two main course objectives (as stated on the syllabus): 1) the ability to think like a microbiologist and 2) the ability to meet the nursing competencies for genetics and genomics. Student responses were characterized, and the number of responses in each category was tallied.
The second part of the attitudinal assessment consisted of IDEA Center Student Ratings of Instruction, using the IDEA Center Diagnostic Form (IDEA Center, 2004). The IDEA diagnostic form provides student feedback to instructors on a number of items. For the purposes of this study, the most important feature is the ability to determine how successfully the course (and instructor) facilitated progress on course learning objectives related to the genetic and genomics competencies (IDEA Center, 2004). The IDEA form allows instructors to pick essential and important objectives from a list of 12 learning objectives. For the Fall 2008 semester, the two essential objectives that were selected were “Developing specific skills, competencies, and points of view needed by professionals in the field most closely related to this course” (IDEA Diagnostic Form, item 24) and “Learning how to find and use resources for answering questions or solving problems” (IDEA Diagnostic Form, item 29). Students responded on a five-point scale (from making no apparent progress [1] to making exceptional progress [5]), from which the IDEA center calculated average scores (on the five-point scale).
RESULTS
The genetics and genomics unit was implemented in three semesters, with slightly different examples and relative emphasis on the technological aspects of establishing genotypes and genetic relationships. In all semesters, content knowledge was assessed by analyzing normalized learning gains on parallel pretest and posttest questions embedded on corresponding course exams.
Content Knowledge Assessment
While the specific assessment questions were revised between semesters, the underlying concepts assessed by the pre- and posttest did not change substantially. In all semesters (Fall 2007, Spring 2008, and Fall 2008), overall normalized learning gains (<g>) for the entire test, as well as learning gains for the biological basis of diversity (Bio) and genetics and genomics technology (Tech), were calculated according to Hake (1998). In the last two semesters (Spring 2008 and Fall 2008), a learning gain for general microbiology (Micro; distinct from genetics and genomics topics) was calculated. The Supplemental Material includes the question classification (Bio, Tech, or Micro), the scoring criteria, and results for representative Bio and Tech questions for the Fall 2008 content test.
Differences between the Pre- and Posttest Scores.
In all three semesters, there was a significant improvement between the average pretest score (ranging from 7.4% to 7.6%) and the average posttest score (ranging from 67.8% to 69.8%; one-tailed paired sample t tests, p < 0.001 for each semester; Table 2). Normalized learning gains (<g>) for each topic (Bio, Micro, and Tech) for each semester (Fall 2007, Spring 2008, and Fall 2008) are also presented in Table 2. In the Fall 2008 semester (Fa08), the individual learning gains followed the trend established in previous semesters: The microbiology learning gain (Micro <g>) was the highest (0.67 in Fall 2008), followed by the biological basis of diversity (Bio <g>; 0.65), with the technology of genetics and genomics (Tech <g>) trailing (0.54; Table 2). A one-way repeated measures ANOVA indicated significant differences overall among subject areas (p < 0.001).
| Semester | Avg. pretest (%) | Avg. posttest (%) | pa | Overall <g> | Micro <g> | Bio <g> | Tech <g> |
|---|---|---|---|---|---|---|---|
| Fall 2007 | 7.6 | 67.8 | <0.001 | 0.65 | ND | 0.69 | 0.57 |
| Spring 2008 | 7.6 | 69.8 | <0.001 | 0.67 | 0.73 | 0.68 | 0.62 |
| Fall 2008 | 7.4 | 68.5 | <0.001 | 0.66 | 0.67 | 0.65 | 0.54 |
Differences between Topics within the Fall 2008 Semester.
Individual contrasts indicated that learning gains for Bio (0.65) and Micro (0.67) were not significantly different (p = 0.421), that Bio normalized learning gains were significantly higher than Tech learning gains (p = 0.002), and that the normalized gain for Micro was higher than the average gains of genetics and genomics (Bio + Tech; p = 0.000). As can be seen in Figure 1, most of the variation among subject areas is due to decreased gains in the Tech scores relative to the other subject areas.
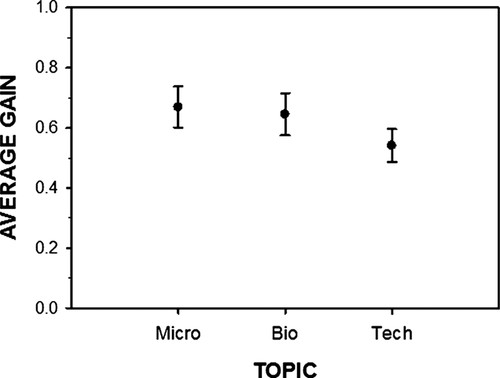
Figure 1. The average normalized learning gains for the three content areas (Micro, Bio, and Tech) with 95% confidence intervals in the Fall 2008 semester are shown.
Differences within Bio and Tech across Semesters.
One-way ANOVAs of Bio and of Tech learning gains indicated no significant differences across semesters (Bio p = 0.690; Tech p = 0.424). Thus minor variations in learning gains for each genetics and genomics topic in each semester (as seen in Table 2) were not significant.
Attitude Assessment
A key question on the end-of-semester anonymous student evaluations asked students for their comments on how well the course helped them meet the two main course objectives (as stated on the syllabus): 1) the ability to think like a microbiologist and 2) the ability to meet the nursing competencies for genetics and genomics. As shown in Table 3, approximately half (n = 22) of the 49 evaluations in the Fall 2008 semester had generically positive comments about the course or the instructor. All of the specific 24 responses (those that specifically mentioned at least one of the two course objectives) indicated confidence in the microbiology course objective. Of these, only 11 clearly indicated confidence in both the microbiology and genetics and genomics objectives. The remaining 13 responses indicated confidence in only the microbiology objective (n = 8) or clear confidence with the microbiology objective and uncertainty with respect to the genetics and genomics competency (n = 5; Table 3).
| Category of comment | No. of comments | |
|---|---|---|
| Generic (e.g., “Course was great” or “Instructor was great”) | 22 | |
| Blank (no response) | 3 | |
| Microbiology objectives were met | 24 | |
| Only microbiology objectives were met | 8 comments | |
| Both objectives (microbiology and genetics/genomics) were met | 11 comments | |
| Microbiology objectives were met; unsure about genetics/genomics | 5 comments | |
| Total responses | 49 | |
The IDEA Center student evaluations, as completed by 50 students in the Fall 2008 semester, showed high ratings of the instructor and the course as well as high ratings on “progress on relevant objectives” (in this case essential objectives; IDEA Center, 1998), indicating that students felt that they made strong progress on the essential objectives of developing relevant skills and competencies necessary to be a practicing nurse and informational literacy (Table 4A).
| A. Overall ratings | Average score | |||
|---|---|---|---|---|
| Overall instructor ratings | 4.9 | |||
| Overall course ratings | 4.8 | |||
| Progress on relevant objectives | 4.6 | |||
| B. Essential objectives | Average score | % 1 or 2 responses | % 4 or 5 responses | |
| “Developing specific skills, competencies, and points of view needed by professionals in the field most closely related to this course.” | 4.5 | 0 | 90 | |
| “Learning how to find and use resources for answering questions or solving problems.” | 4.6 | 2 | 96 |
For the essential objective of developing relevant skills and competencies (“Developing specific skills, competencies, and points of view needed by professionals in the field most closely related to this course”; IDEA Diagnostic Form, item 24), the average score was 4.5 (on a five-point scale, with 1 being low and 5 being high), with 90% of all responses being a 4 or a 5 (Table 4B). For the essential objective of informational literacy (“Learning how to find and use resources for answering questions or solving problems”; IDEA Diagnostic Form, item 29), the average score was 4.6 (on a five-point scale, with 1 being low and 5 being high), with 96% of responses being a 4 or a 5 (Table 4B).
DISCUSSION
The critical importance of genetics and genomics in personalized and genomic medicine is apparent (Hoopes, 2008; Feero et al., 2010; Hamburg and Collins, 2010; Varmus, 2010). The new genetics and genomics competencies for nurses recognize the importance of this emerging field but present a challenge, as they require education and professional development for both preservice nursing students and practicing nurses (Jenkins et al., 2001). One strategy that has been put forth is to distribute concepts critical to genetics and genomics throughout the prenursing basic science curriculum (Guttmacher et al., 2007). These courses often include introductory biology, human physiology, human anatomy (with lab), and microbiology (with lab). This project was designed to test the hypothesis that concepts relevant to the nursing genetics and genomics competencies can be taught and effectively learned in a prenursing microbiology lecture course.
Although many of the competencies are clearly specific for human genetics and genomics (e.g., “Constructs a pedigree from collected family history information using standardized symbols and terminology” or “Identifies clients who may benefit from specific genetic and genomic information and/or services based on assessment data”), many are broad enough that they can be reinforced with any model system that addresses genotype/phenotype relationships in the context of diagnosis and selection of targeted or personalized therapy (e.g., “Demonstrates an understanding of the relationship of genetics and genomics to health, prevention, screening, diagnostics, prognostics, selection of treatment and monitoring of treatment effectiveness” or “Identifies credible, accurate, appropriate, and current genetic and genomic information, resources, services, and/or technologies specific to given clients”; Consensus Panel on Genetic/Genomic Competencies for Nurses, 2006). Such concepts are at least theoretically addressable in a microbiology course, when considering genotype/phenotype relationships and the implications of genotypic (and corresponding phenotypic) diversity in the choice of appropriate antibacterial or antiviral therapy.
The main question being explored here is whether students can learn genetics and genomics content in the context of a microbiology lecture course. Based on pre- and posttest scores and normalized learning gains, the short answer appears to be that, yes, they can. The posttest scores are significantly higher than pretest scores for all the genetics and genomics topics (Table 2), and the average normalized learning gains are comparable to improved gains seen following the adoption of interactive learning strategies into a course (Hake, 1998; Knight and Wood, 2005). In a study of thousands of physics students, Hake (1998) found normalized learning gains ranging between 0.34 and 0.69 (average = 0.48) for courses taught with interactive engagement, as compared with normalized gains averaging 0.23 for courses taught by traditional methods. Similarly, Knight and Wood (2005) found normalized gains of approximately 0.62 in semesters with interactive engagement, in contrast to normalized gains of 0.46 in semesters using traditional instruction. In fact, based on the Fall 2008 data, the average normalized learning gain for Micro (0.67) is indistinguishable from that for the genetics subtopic of Bio (0.65; ANOVA, p = 0.421). This is encouraging, as it suggests that aspects of genetics and genomics can be taught in a microbiology course as effectively as general microbiology topics. However, the learning gains for genetics and genomics technology (Tech) were significantly lower than the gains for Bio (ANOVA, p = 0.002). In all three semesters, Tech normalized learning gains were lower than Bio normalized learning gains (Table 2). This suggests that the technology used to assess genetic diversity and make genetics-based diagnoses may be more difficult for students to understand than other topics in genetics. This is not surprising, as many of the technologies (e.g., DNA profiling and PCR) are somewhat abstract and often relegated to a chapter on biotechnology in microbiology textbooks rather than being discussed and integrated in a diagnostic and therapeutic context. If the community agrees that better familiarity with genetics and genomics technology is indeed critical for nurses, then we will need to assess strategies to accomplish this, including developing and assessing specific laboratory (hands-on) exercises.
The learning gains data thus suggest that students can indeed learn genetics and genomics content in an allied-health microbiology course. According to the attitudinal assessments, students responded equally favorably to the two IDEA Diagnostic items (general skills and competencies and literacy skills), suggesting that they felt that the course prepared them for professional practice (Table 4). Reviewing the open-ended responses to a specific inquiry about meeting course objectives (thinking like a microbiologist and meeting genetics and genomics competencies), it appears that students are perhaps less assured about their confidence in genetics and genomics competencies than in thinking like a microbiologist (Table 3). This may reflect that they are genuinely less confident about genetics and genomics and/or that they have a less concrete sense of the applications of genetics and genomics in a microbiology or nursing context. It also raises the question of how well genetics learned in a microbiological context will be transferred to a purely human context.
While the results of this project are positive in terms of learning gains achieved for genetics and genomics content taught in an allied-health microbiology course, they suggest that genetics technology was not learned as well and that students are less confident about the applications of and their abilities in genetics and genomics. This suggests a number of lines of investigation. One possibility is to reinforce genetics technology in a carefully coordinated set of lab exercises. Another possibility is to integrate genetics and genomics into a variety of courses in the prenursing science curriculum. For example, many metabolic disorders (inborn errors of metabolism) are the result of inheriting particular alleles and provide strong reinforcement of genetics and genomics in the context of a physiology course. It may be that teaching genetics and genomics in an explicitly human context will be even more effective than introducing it in the context of infectious disease. But this is a question that requires additional investigation and consensus in the field about key aspects of genetics and genomics for nurses. This report is intended to start the conversation and emphasize the critical importance of evidence-based decisions in genetics and genomics education for preservice allied-health students.
ACKNOWLEDGMENTS
This work was funded by NMSU ADVANCE, the College of Arts and Sciences at NMSU (start-up funds), and the Department of Biology at NMSU (start-up funds). The assistance of William Boecklen with statistical expertise and manuscript review is gratefully acknowledged, as is the support provided by Tara Gray of the NMSU Teaching Academy for the IDEA evaluations and manuscript review. The assistance of C. Brad Shuster with the preparation of figures and manuscript review was invaluable to the completion of this work.
This material is based upon work supported by the National Science Foundation under grant NSF-0123690. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.



