Using Long-term Time-lapse Imaging of Mammalian Cell Cycle Progression for Laboratory Instruction and Analysis
INTRODUCTION
Video Microscopy Movies as a Teaching Tool for Cell Biology Laboratories
Cinemicrography — the capture of moving cellular sequences through the microscope — has been influential in revealing the dynamic nature of cellular behavior (reviewed in Rose, 1963). Initially, cinemicrographic imaging was done using silver halide-based film, and these movies were instrumental in demonstrating the dramatic behavior of cell motility (Bajer and Bajer, 1972). Over the last 25 years, cell biology has benefited from improvements in electronic imaging technologies that have largely replaced silver-based film recordings. During the 1980s the use of analog video technology greatly expanded the use of light microscopy as an analytic tool (Inoué, 1986; Salmon, 1995). Over the last several years, the use of analog video-capture systems has been largely replaced by computer-based digital image-capture systems (Inoué and Spring, 1997; Sluder and Wolf, 2003). With their high quantum efficiency, low-noise characteristics, and ease of use, imaging systems for digital micrography have revolutionized the study of the dynamic events of cell biology.
The recent advances in imaging technology have been coupled to improvements in photochemistry/photobiology, with the development of sophisticated molecular probes that have allowed the visualization of discrete molecules within living cells (Lippincott-Schwartz et al., 2001; Rieder and Khodjakov, 2003). The technological advances in these probes have allowed sophisticated molecular interactions to be studied at the level of the individual cell (Cardullo and Parpura, 2003). A major advance in the bioimaging field has been the discovery of green fluorescent protein (GFP; reviewed in Lippincott-Schwartz and Patterson, 2003), which allows individual proteins to be visualized and imaged. Chimeras made from any gene of interest coupled to GFP — and to genetically engineered chromatic variations of GFP — can be easily introduced into cultured cells as well as genetically tractable organisms such as yeast, flies, worms, and fish (Zhang et al., 2002; Haraguchi, 2002). More often than not, these GFP fusions retain their native biological activity while becoming fluorescently tagged. This technique has become a powerful tool for the investigation of the dynamics of individual proteins at the level of the single cell.
In general, time-lapse digital microscopy provides an important tool for the instruction of students in a cell biology laboratory. The resultant image sequences portray the true nature of cellular processes and make it much easier for students to grasp the dynamics of these events. This is particularly true for movies of GFP-tagged proteins: they provide a direct way to assay for molecular interactions within the cell (Straight et al., 1996). But cinemicrographic sequences are not just pretty pictures; digital micrography sequences provide a wealth of analytical information that can be readily used by students in a laboratory course, even if equipped with only basic computer tools. Each student or group of students can be given the image file(s) and use inexpensive/free software to conduct very informative image analysis experiments (see analytical software tools developed by the Laboratory for Optical and Computational Instrumentation, LOCI, at the University of Wisconsin: http://www.loci.wisc.edu/ and the freeware program Image-J, developed by the NIH: http://rsb.info.nih.gov/ij/). Thus, a library of time-lapse image data is essential not only for demonstrating cellular processes but also to provide the basis for data interpretation and analysis by students in a laboratory course. It is ultimately these analytical skills that students need to acquire as they prepare for careers in research.
Although some undergraduate laboratories are equipped to conduct cutting-edge, live-cell, time-lapse imaging, many are not. Setting up an imaging system to conduct these experiments can be problematic. The sophisticated equipment needed, although readily available, also comes with a substantial price tag. In addition, there is a certain amount of expertise necessary to conduct live-cell biology and maintain the imaging equipment. The use of simple laboratory organisms, such as the early embryos of marine invertebrates, fruit flies, or nematodes, can often circumvent these difficulties. A wealth of information can be obtained using invertebrate cells, and their culture and maintenance under the microscope are greatly simplified (Morris et al., 2004). These organisms are not only suited for teaching labs but also provide ideal experimental systems to be used with undergraduate research students as well as serving as the experimental organisms for big research labs in medical schools. However, if the laboratory is to use cultured mammalian cells for live-cell experiments, then the experimental needs and technical expertise increase (see Goldman and Spector, 2005, for detailed methods on the imaging of living cells). An innovative solution to this problem is for the cell biology research field to provide digital image sequences as part of an increasingly available archive that can be utilized as part of a laboratory course. Cell Biology Education (CBE) has already begun this endeavor with the publication of the Video Views and Reviews series (see Watters, 2003, and in this issue), which reviews annotated video files that exist in the Journal of Cell Biology collection and elsewhere. The use of movies published as part of research articles can be very beneficial to students, but not all great movies end up as part of peer-reviewed publications. In addition, movies submitted and archived by research journals are not always designed with the laboratory student in mind. The scope of the present article is to begin to provide novel, unpublished movies — along with a description of materials/methods, results, and discussion of the work — specifically for the CBE audience. If these movies become available to the cell biology community, even modest cell biology laboratory programs can begin to utilize this ever-increasing library of data to both demonstrate the movement of the cell and provide live-cell data to analyze in the laboratory.
The Role of Video Microscopy in Studying Cell Division
One of the more dramatic cellular events is mitosis, the division of sister chromatids into two daughter cells. Mitosis has been extensively studied in a variety of organisms, both morphologically and at the molecular level (Scholey et al., 2003). Importantly, the study of mitosis has been driven by the use of the light microscope (elegantly reviewed in Rieder and Khodjakov, 2003). Static (i.e., fixed-cell) studies of cell division revealed the structure of mitotic cells and, in particular, the organization of the mitotic spindle (Sluder et al., 2004). However, many key events during mitosis occur only transiently; the spindle undergoes dramatic morphological changes over the period of an hour or less, and the significance of these events is often lost in fixed time-point samples. Many of the important events in cell division can be missed if not imaged continuously. Video microscopy also provides the ability to measure the duration of key events and to examine the dynamic redistribution of structural and regulatory proteins as the spindle assembles, aligns the chromosomes, and segregates the sister chromatids at anaphase. Importantly, the repeated, direct measurement of cell division events by time-lapse microscopy ensures that critical observations are believed and substantiated rather than being dismissed as an artifact of fixation or as a one-time “bizarre cell behavior.”
With the advent of time-lapse imaging, researchers have begun to directly examine and test specific models of how the spindle assembles and mediates proper chromosome segregation. Video microscopy has been fundamental in understanding these dynamic events, including the behavior of the spindle fibers (microtubules), movement of chromosomes, and the cell cycle feedback mechanisms that monitor proper attachment of the chromosomes to the spindle (Rudner and Murray, 1996). Coupling video microscopy with fluorescently labeled proteins, such as tubulin, actin, and chromosome passenger proteins, researchers have revealed many of the molecular mechanisms that control cell division (Wang, 2001; Wadsworth and Khodjakov, 2004). A recent and fascinating development has been the generation of stable cell lines expressing α-tubulin coupled to GFP (Rusan et al., 2001). This has allowed for the examination of the dynamic microtubule events in living, dividing cells, including spindle assembly and mitosis, and mitotic exit (Rusan et al., 2002; Tulu et al., 2003; Rusan and Wadsworth, 2005).
The interest in our lab lies in understanding the regulation of mitotic spindle assembly and how cells exit from mitosis. Because our experimental manipulations often delay mitosis for several hours, we have developed methods to image cells over long periods of time, on the order of days. To directly examine the dynamics of spindle assembly in living cells, we have generated a line of epithelial cells constitutively expressing α-tubulin-GFP and use a new slide preparation that allows for imaging of cells over the period of several days (Hinchcliffe et al., 2001; Sluder et al., 2005). These time-lapse images can reveal much about the dynamic behavior of cells as they transit over multiple division cycles and, as a result, should provide interesting information for students in a laboratory course.
MATERIALS AND METHODS
The methods presented here will allow instructors and students to critically analyze how the experiments were done in order to interpret the results. In addition, an extensive description of the imaging techniques used is presented, should laboratories wish to conduct these experiments.
Establishing the Cell Line
BSC-1 (monkey kidney epithelial) cells were obtained from the American Tissue Culture Collection (ATCC; Manassas, VA). These cells were maintained in Dulbecco's Modified Eagle Media (DMEM; Sigma, St. Louis, MO), pH 7.2, with 10% fetal bovine serum and penicillin/streptomycin. These cells were grown on tissue culture plates in an incubator at 37°C and 10% CO2.
BSC-1 cells were transiently transfected with human α-tubulin coupled to EGFP (Invitrogen, Carlsbad, CA), and a stable cell line was isolated, essentially as described (Rusan et al., 2001). For transfection, cells were plated out at high density, and the expression plasmid encoding human α-tubulin coupled to GFP was transfected using Fugene 6 (Roche, Indianapolis, IN). Transfected cells were then subjected to selection with G418 (Sigma), which causes nonexpressing cells to die. Individual cell colonies (clones) were withdrawn from the culture using a 10-ml pipetter and a yellow tip, grown in culture dishes, and then subjected to limited dilution to isolate single colonies that expressed the GFP-fusion protein. The resultant clones were then screened by fluorescence microscopy to identify cells expressing low levels of GFP that did not interfere with normal cell function. Final clones (BSC-1α -tubulin GFP) were generated from a colony of cells that was derived from a single cell plated into 6-well dishes on a feeder layer of PtK1 cells (see Khodjakov and Rieder, 1999). Positive clones were stored in liquid nitrogen until used.
Imaging Cells
For time-lapse imaging, BSC-1 α-tubulin GFP cells were plated onto biocleaned glass coverslips with a wax pencil line drawn 5 mm from one edge (see Sluder et al., 1999, for the biocleaning protocol). Cells were cultured on coverslips in imaging medium (DMEM w/o phenol red, containing 12 mM Hepes, pH 7.2, and 10% fetal bovine serum: Sigma). Cells on coverslips were then assembled onto aluminum support slides, as described (Hinchcliffe et al., 2001; Sluder et al., 2005). Essentially, these support slides are modified Kiehart chambers, originally designed for the microinjection of sea urchin eggs (for a diagram and detailed description of these chambers, see Kiehart, 1982; also see Sluder et al., 1999). To assemble these chambers, coverslips were assembled onto the top surface of the chamber using silicon grease (Dow Corning, Midland, MI), and another coverslip (without cells) with a matching wax pencil line was assembled onto the bottom surface. The two wax pencil lines run parallel to the front edge of the chamber, which remains open. The chamber is then filled with 100 μl FC-43 fluorocarbon oil (Minnesota Mining and Manufacturing, St. Paul, MN), and then 200 μl imaging medium was added, using a syringe and needle. The gap between the upper and lower wax pencil lines and the outer (i.e., front) coverslip edge was capped with mineral oil (to prevent drying). Finally, the top surface of the coverslip (with cells on it) was washed with 378C dH2O, by dripping the water onto the surface with a squeeze bottle and coordinately sucking the water off with a Pasteur pipette attached to a vacuum trap (see Sluder et al., 1999).
Time-lapse Digital Microscopy
Cells cultured in imaging chambers were imaged by wide-field microscopy using either phase-contrast or fluorescence optics. The phase-contrast images and the fluorescence images were taken using the “high-tech” microscope (see below). We also describe the “low-tech” microscope, which is a more affordable solution for live-cell imaging.
The “high tech” scopes are based on a Leica DM RXA2 motorized, upright microscope stand (Leica Microsystems, Bannockburn, IL). These microscopes are equipped with fluorescence, phase, and differential interference optics. The stands are enclosed in a custom-made Plexiglas box and heated with an industrial shrink-wrap heat gun (Master Mite, Master Appliances, Racine, WI). The temperature in the box is maintained at 37°C using a proportional heat controller (CN9000A, Omega Electronics, Stamford, CT) coupled to the heat gun. Images are taken using a Leica Plan Apo 20×/0.7 NA CORR multi-immersion objective (using glycerol as the immersion medium) or a Leica Plan Apo 63×/1.3 NA 37°C glycerol immersion objective. In addition, the images are projected onto the face of the CCD camera by means of empty magnification (discussed in Hinsch, 2003) via a Bertrand lens (maximum ×1.6 magnification) and a Leica Vario-tube zoom lens (×0.33-1.6 magnification). The GFP was excited using an Hg arc lamp, using a GFP narrow band-pass filter set, and infrared radiation was filtered from the light source by a hot mirror (both from Chroma Technology, VT). To prevent photobleaching during the experiment, the fluorescence illumination pathway was shuttered with a filter wheel (Ludl Inc., Hawthorne, NY). Images are captured using a Hamamatsu ORCA-ER Digital CCD camera (Hamamatsu, East Bridgewater, NJ). The digital signal was sent into a Matrox Millennium image-capture card mounted in a PCI slot of a personal computer. The time-lapse images were generated using Simple PCI imaging software (Compix, Brandywine, PA). Time-lapse image sequences were converted from the Compix cxd file format to audio-video interleave (AVI) movies.
The low-tech microscope consists of a 25-year-old Zeiss Universal microscope (Carl Zeiss, Thornwood, NY) with finite (i.e., 160 mm) tube optics enclosed in a cardboard box and heated with an industrial shrink-wrap heat gun (Master Mite, Master Appliances, Racine, WI). The temperature in the box was maintained at 37°C using a proportional heat controller (CN9000A, Omega Electronics, Stamford, CT) coupled to the heat gun. Cells are imaged using a Zeiss Neofluor 20×/0.5 NA Phase objective. The light from the 12-W halogen lamp is filtered by a green filter (546 ± 40 nm bandpass), and a KG-5 heat-cut filter (both from Chroma, Burlington, VT). Images are captured using an analog Hamamatsu C2400 charge-coupled device (CCD) camera controlled by a Hamamatsu Argus 20 image processor (Hamamatsu, East Bridgewater, NJ). The analog video signal was sent through a RasterScope (Dage/MTI, Michigan City, IN) to ensure that the maximum range of the signal was used (see Hinchcliffe and Sluder, 2003) and then fed into a MiroVideo DC30 video capture card (Pinnacle Systems, Sunnyvale, CA) mounted in a PCI slot of a personal computer. Time-lapse images are captured using Adobe Premiere 4.0 (Adobe, Mountain View, CA) and saved to a computer's hard drive as AVI files using MJPEG hardware compression/decompression driven by the DC30 MJEG capture card. This low-tech setup can be recreated by using any phase-contrast microscope and a video-rate camera system (video tube camera, or video-rate CCD). This substantially reduces the cost, though such cameras have fairly high noise and may not be suitable for fluorescence imaging.
AVI movies generated on the “low-tech” scope must be recompressed in Adobe Premiere, using a software compression/decompression routine (known as a codec), allowing them to be played on most personal computers. The choice of compression codecs for digital movies is best determined empirically for a given set of imaging conditions (discussed in Hinchcliffe, 2003).
RESULTS
Long-term Imaging in Phase Contrast
We continuously imaged BSC-1 cells for 67 h by time-lapse microscopy using phase-contrast optics (Figure 1 and Supplemental Movie 1). The time stamp in the upper-left corner gives hours and minutes. Note that the counter resets after ∼52 h as we stopped recording, checked the movies, and then restarted. Initially there are two cells, which, given their morphology, probably arose from a single mother cell in the previous cell cycle. The motion picture sequence shows these two cells each undergoing several rounds of division. The first round of mitosis is relatively synchronous between the two cells, with both entering M-phase within 15 min of each other and dividing to yield four cells (Figure 1B). As the sequence proceeds, the cells continue to divide. Each cell divides at least twice, and some of the cells in the colony undergo four complete rounds of division. Note that the colony of cells — originally derived from two cells — grows to contain 24 cells, 23 of which are derived from the initial two cells, and one cell that migrates into the frame from below and makes a connection with the initial colony (Figure 1).
Figure 1. Frames from Supplemental Movie 1, showing BSC-1 cells undergoing repeated cell division cycles. The two initial cells in frame A divide repeatedly over 67 h. Note the position of the centrosome can be imaged in the cells by the position of the mass of granules near the nucleus (A, arrow). Cells undergoing mitosis can also be visualized (arrows in C and F). Elapsed time in hours and minutes is shown in the lower-left corner. Phase contrast. Bar = 15 μm.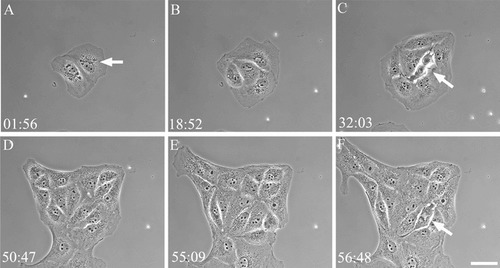
Phase contrast can reveal important information about the division cycle of the cell. Using this contrast mode, we can image the cell outline, the position and movement of granules, and the nucleus and nucleolus (dark circles within the nucleus; Figure 1). Interestingly, using phase-contrast microscopy, we have observed that BSC-1 cells undergo nuclear rotation before the onset of mitosis, and this can be seen in the time-lapse sequences presented (Supplemental Movie 1). The function for this rotation is not known. These phase-contrast images also reveal that the nuclear envelope breaks down before or coincident with the rounding up of the cells. This reveals how cells coordinate the molecular events of mitotic onset (activation of the cyclin-dependent kinases) with the change in cell morphology (rounding up: see Hinchcliffe et al., 1999).
Long-term Imaging Using Fluorescence Optics
BSC-1 α-tubulin GFP cells were continuously imaged using fluorescence optics for 188 h (Figure 2 and Supplemental Movie 2). When fluorescence coupled with expression ofα -tubulin GFP is used, this view of cell cycle progression and cell division is entirely different from that seen with phase contrast in Figure 1. There is little detail of the nuclei; in fluorescence the nuclei appear as dark circles because they exclude the tubulin fluorescence found in the cytoplasm. At the relatively low resolution/low magnification of these images, a prominent feature seen in these cells is the major microtubule-organizing center (MTOC) within each cell, which represents the position of the centrosome (Figure 2B, arrow). This MTOC appears as a bright fluorescent region near the nucleus. If the movie is presented by slowly stepping through the cine sequence, the viewer can see that the centrosome duplicates to yield the two poles of the mitotic spindle (Figure 2I, arrows), though these organelles often have not separated sufficiently before spindle formation to be visible in all cells.
In the phase-contrast sequence, the chromosomes are clearly visible during M phase; in the fluorescence images, the major mitotic feature is the microtubule-based spindle. When the movie is played at a fast frame rate (rapid playback), these spindles appear like popcorn, forming as bright structures and then quickly fading as the cells exit mitosis. This observation drives home the notion that the cell undergoes rapid and extensive remodeling of its cytoskeleton both during the transition into M phase and following its exit from mitosis. Another key observation that can be made from watching these movies is the appearance of the midbody, which connects the two daughter cells following cytokinesis (see Figure 2K, arrow). These structures persist after the cells have completed mitosis. The midbodies are eventually excised and lost, and this process is clearly visible in the video sequences.
DISCUSSION
The long-term time-lapse imaging of cells undergoing multiple cell cycles provides an important view into the process of cell division and how this process is integral for the life of the cell. Because BSC-1 cells are epithelial, they seek to form continuous sheets of cells. This can clearly be seen in both video sequences, as dividing cells form separate colonies, migrate together, and form cell-cell attachments. Because these video sequences span several days (and multiple cell cycles), observing these movies reinforces the idea of the dynamic nature of the division cycle and how this results in cells interacting as they develop.
Figure 2. Frames from Supplemental Movie 2, showing BSC-1 cells undergoing repeated cell division cycles. The colony of 15 cells in the center of frame A divides repeatedly over 188 h. Note the position of the centrosome can be imaged in each cell as a bright focus adjacent to the nucleus (B, arrow). The single interphase centrosome duplicates to provide two spindle poles; this can briefly be seen in select frames (for example, I, arrows). As the colonies continue to grow, cells that have assembled metaphase spindles can easily be visualized (arrows in frames C, E, F, and L). Cells in anaphase, where the chromosomes segregate, can also be seen in these movies (K). Select frames from these movies also reveal the position of the midbody, which assembles as the cell cleaves into two, then persists into the next cell cycle (K, arrow). Elapsed time in hours and minutes is shown in the lower-left corner. Fluorescence contrast. Bar = 35 μm.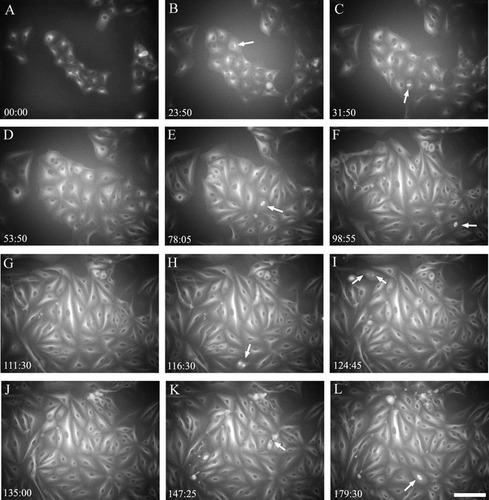
Observing movies of repeated cell division also can provide a wealth of simple data that can be analyzed by laboratory students. These data are truly multiscale. First, the sequences offer the opportunity to analyze cellular events that can be addressed on a cell-by-cell basis: questions such as the time each cell takes to complete mitosis, the duration of interphase between successive divisions of daughter cells, and the lifetime of the midbody before excision can be directly addressed using these data. The behavior of the cells in these movies can also be treated as a population: the analysis and modeling of the specification of the division planes within the population, the change in cell cycle times as the sequence progresses, any patterns in epithelial growth, and frequency of mitotic mistake (there are a few, evident to the careful observer) can all reveal important and novel aspects of how mammalian cells function in culture.
There are also limitations to analyzing these movies, and these limitations should provide the basis for a discussion with students about how experiments are designed and how data are handled. Students may find that the frame rate (one frame per 3 or 5 min) is not sufficient to be able to critically analyze certain events. An obvious solution would be to increase the frame rate. But this solution raises a central question in live-cell imaging: what will the effects on the cells be? Increasing framing of fluorescent excitation will invariably lead to photobleaching and/or photodamage to the cells. There will also be an increase in file size, and how to deal with these“ real-life” concerns can be addressed as part of a laboratory discussion.
Individual frames from these movies can be exported as TIFF files using Adobe Premiere. Quantitative analysis can then be conducted on these TIFF files using the freeware program Image-J ( http://rsb.info.nih.gov/ij/). Thus, changes in fluorescence intensity over time can be measured for selected frames. However, it is important to recognize that the original video files were compressed, and any quantitative analysis will be on compressed data. There are certainly more questions that can be asked using these data, and it will be left up to the students and instructors of laboratory classes to begin to do so.
FOOTNOTES
Monitoring Editor: Dennis Liu
ACKNOWLEDGMENTS
The long-term time-lapse methods described in this paper were originally developed while the author was an American Cancer Society Post-Doctoral Fellow (Massachusetts Affiliate) in the laboratory of Kip Sluder, and I would like to thank Kip, along with Rick Miller, for teaching me the finer points of live-cell imaging. I would also like to thank my students for their continuing interest in using light microscopy to study cell biology. The author is a Research Scholar of the American Cancer Society and is supported by ACS RSG CCG-104915, and NIH RO1 GM07275.