Pathways over Time: Functional Genomics Research in an Introductory Laboratory Course
Abstract
National reports have called for the introduction of research experiences throughout the undergraduate curriculum, but practical implementation at many institutions faces challenges associated with sustainability, cost, and large student populations. We describe a novel course-based undergraduate research experience (CURE) that introduces introductory-level students to research in functional genomics in a 3-credit, multisection laboratory class. In the Pathways over Time class project, students study the functional conservation of the methionine biosynthetic pathway between divergent yeast species. Over the five semesters described in this study, students (N = 793) showed statistically significant and sizable growth in content knowledge (d = 1.85) and in self-reported research methods skills (d = 0.65), experimental design, oral and written communication, database use, and collaboration. Statistical analyses indicated that content knowledge growth was larger for underrepresented minority students and that growth in content knowledge, but not research skills, varied by course section. Our findings add to the growing body of evidence that CUREs can support the scientific development of large numbers of students with diverse characteristics. The Pathways over Time project is designed to be sustainable and readily adapted to other institutional settings.
INTRODUCTION
A series of national reports have called for the incorporation of undergraduate research experiences (UREs) throughout the undergraduate curriculum (National Research Council, 2003; American Association for the Advancement of Science [AAAS], 2011; National Academies of Science, Engineering, and Medicine, 2015), based on their documented benefits to students. Students participating in UREs report the acquisition of scientific skills; increased self-identification as scientists; increased communication and collaboration skills; and greater persistence in science, technology, engineering, and mathematics (STEM) disciplines (Seymour et al., 2004; Hunter et al., 2006; Lopatto, 2007; Harrison et al., 2011; Eagan et al., 2013). These gains are particularly significant for students from underrepresented minority (URM) groups, who typically are less likely to persist as majors in STEM disciplines (Lopatto, 2007; Jones et al., 2010; Chen and Soldner, 2013; Hernandez et al., 2013). Traditionally, UREs have been independent research projects in which students pursue an authentic research question related to the research interests of a faculty mentor. The numbers of students who are able to participate in independent UREs are typically limited, however, especially at universities with large undergraduate enrollments and at community colleges.
Classroom-based undergraduate research experiences (CUREs) provide an alternative mechanism to engage larger numbers of students in research, often with student outcomes similar to those of traditional UREs (Shaffer et al., 2010; Harrison et al., 2011; Kloser et al., 2011; Harvey et al., 2014; Jordan et al., 2014; Corwin et al., 2015a; Shapiro et al., 2015). Recognizing the potential of CUREs to increase both the number and diversity of STEM graduates, the Presidential Council of Advisors on Science and Technology (2012) has called for the introduction of discovery-based research classes for all students within their first 2 years of college. Bangera and Brownell (2014) have suggested that the incorporation of CUREs into core curricula could ensure that all students participate in research at the undergraduate level.
Laboratory courses are natural settings for undergraduate research, because it is here that students practice how to think and act like scientists. The introduction of authentic research experiences into introductory laboratories, however, can be challenging. In a recent survey of 279 biology faculty, most of whom served as course instructors or course directors, respondents identified multiple barriers to implementation. These barriers, which varied by institution type, included large class sizes, multiple course sections, cost, alignment with lecture classes, and student underpreparation (Spell et al., 2014). In general, fewer research activities were incorporated into biology laboratory courses at 2-year colleges, and the fraction of course time devoted to research activities decreased as the number of sections increased. Interestingly, faculty definitions of authentic research experiences in this same survey fell into two distinct classes. The predominant class of responses emphasized elements of the scientific process, such as hypothesis generation, experimental design, data collection/analysis and presentation. A smaller number of respondents emphasized the novelty of research questions as the defining characteristic of research experiences.
A group of educators with expertise in CURE instruction recently identified common design features of CUREs that have been useful for their assessment (Auchincloss et al., 2014; Corwin et al., 2015b): the use of scientific practices, discovery, broader relevance, collaboration, and iteration. Some of these features are not unique to CUREs, and existing CUREs fall along a continuum within these categories. For example, CUREs may incorporate some structured or guided-inquiry activities to teach specific skills, even though the overall student experience is built around an authentic research question whose answer is unknown (Brownell and Kloser, 2015).
Although the most common CUREs are still small laboratory courses in which advanced students work on projects related to a faculty member’s research, new CURE models have been developed for larger numbers of introductory students. Individual faculty members have developed introductory CUREs in ecology (Kloser et al., 2011) and genetics (Burnette and Wessler, 2013) that were associated with gains in student self-efficacy and research skills similar to those reported for traditional UREs. Institutional support can help to promote the development of CUREs for introductory students. The Freshman Research Initiative (FRI) at the University of Texas provides a notable example in which ∼900 freshmen participate in ∼30 different faculty-directed research streams. Implementation of the FRI requires significant institutional resources, including dedicated PhD-level research educators and graduate teaching assistants (GTAs) who supervise the undergraduate projects, as well as lab spaces for the different research streams. Students who completed the three-semester FRI program were significantly more likely than their demographically matched non-FRI peers to complete a STEM degree and to graduate (Rodenbusch et al., 2016).
Collaborative CUREs provide another model in which students at institutions with limited research resources are able to participate in an authentic research project developed by a leading investigator at a central site. In this distributed model, the central site provides curricular materials and research support to students at remote sites, who work in parallel under the supervision of a local faculty member who has received training in the research project. Students submit their data to the central site, where the data are aggregated and interpreted for research publications. Several projects have demonstrated the success of this model. The Undergraduate Genomics Research Initiative involved thousands of students in the sequencing and annotation of Ammonifex degensii, a thermophilic bacterium (Kerfeld and Simon, 2007). In the widely adopted SEA-PHAGE project (https://seaphages.org), classes of introductory college students isolate novel bacteriophages from local environmental samples and annotate the phage DNA sequence information, which is provided by the central site. The aggregated student results have provided novel insights into viral diversity and generated multiple publications, with measured student outcomes similar to those of traditional UREs (Hatfull et al., 2006; Jordan et al., 2014; Pope et al., 2015). In the Genomics Education Partnership (https://gep.wustl.edu), students use bioinformatics tools to annotate sequence information for Drosophila “dot” chromosomes. The aggregated student data has contributed to new models for the diversification of Drosophila genomes (Slawson et al., 2006; Leung et al., 2015). Genomics Education Partnership faculty attest to the importance of the central support system in the sustainability and implementation of the program (Lopatto et al., 2014).
The development of CUREs for multisection laboratory core courses presents challenges related to the nature of the research question, sustainability, equity, and cost. These courses are typically led by a PhD-level instructor(s) who oversees a support staff that may include technicians, GTAs, and undergraduate assistants. At many universities, the composition and expertise of this instructional team can change from one course offering to the next. To transform these introductory laboratories into CUREs, some departments have developed course research projects that are not linked to a particular faculty member’s research. For example, students in a molecular biology laboratory course at Brandeis University explore the effects of amino acid substitutions on the stability of a lens crystallin. Assessment data suggest that the course effectively teaches protein structure–function relationships while promoting critical thinking skills and interest in biology (Treacy et al., 2011). Students at Stanford University test the ability of p53 tumor–associated variants to activate transcription of p53-responsive reporter genes in a Saccharomyces cerevisiae model. Data collected concerning this project suggest that students’ scientific thinking becomes more expert-like by the end of the semester (Brownell et al., 2015; Hekmat-Scafe et al., 2016).
Investigations in Molecular Cell Biology or BI204 (since renamed BIOL2040), the subject of this study, is a novel 3-credit core course that immerses introductory students in a semester-long project involving functional genomics. The course replaced two previous 1-credit laboratory classes in genetics and molecular cell biology. Classes meet twice a week for 3-hour sessions. This biweekly format was chosen for several reasons. More frequent class meetings may promote GTA–student relationships (Kendall and Schussler, 2013) and build a sense of community, which may increase retention of students in STEM majors (Jones et al., 2010). The format also provides students more time to work on their research projects, a factor that positively affects student outcomes in CUREs (Shaffer et al., 2014; Linn et al., 2015). Finally, the format eliminates problems commonly associated with aligning lecture and lab content (Spell et al., 2014). As reported in the research portion of this article, assessment data show consistent student gains in both research skills and content knowledge during participation in the course.
COURSE DESIGN
Pathways over Time: A Research Project in Functional Genomics
Functional genomics was chosen as the project research theme because whole-genome sequencing (WGS) projects are providing massive amounts of data that can be used to shape authentic research questions suitable for investigation by undergraduates. WGS projects use computational methods to identify genes by their homology to known genes, but functional testing is not part of the process. Undergraduate research projects can help to fill the gap between predicted and demonstrated function. In the Pathways over Time project model (Figure 1), students study the functional conservation of proteins in a multi-enzyme pathway. The project is designed to incorporate the core concepts for biological literacy identified in the Vision and Change report (AAAS, 2011): evolution, structure–function relationships, information transfer, pathways, and systems. Three basic decisions dictate the general project design, which can be tailored to different settings and adapted as data accumulate. Instructors and/or students select 1) a multi-gene system in which mutant phenotypes can be identified in a genetic screen, 2) a genetically tractable organism that that can be used to analyze protein function in complementation assays, and 3) an evolutionarily divergent organism whose genome sequence contains orthologous genes to test for function in the genetic model. The project was designed to be flexible, recognizing that course instructors may want to design a research question that aligns with their particular interests and research experience.
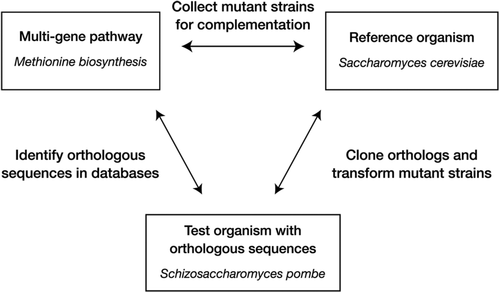
FIGURE 1. Pathways over Time project design. Project design involves three elements: a multi-gene pathway, a reference organism, and a test organism that serves as the source of orthologous sequences for cross-species complementation. The course project described in this study centered on the conservation of genes involved in methionine synthesis between S. cerevisiae, the reference organism, and the fission yeast S. pombe.
Over the five semesters documented in this study, Boston College students studied the functional conservation of the enzymes involved in methionine (Met) biosynthesis (Thomas and Surdin-Kerjan, 1997) between the budding yeast, Saccharomyces cerevisiae, and the fission yeast, Schizosaccharomyces pombe (Hebert et al., 2011). Methionine is an essential cellular molecule in all domains of life because of its roles in protein synthesis and methylation reactions, and the genes involved in Met synthesis show interesting evolutionary plasticity (Gophna et al., 2005). The pathway involved in Met synthesis in S. cerevisiae (Figure 2) is easy for students to visualize, because Met is one of only two sulfur-containing amino acids.
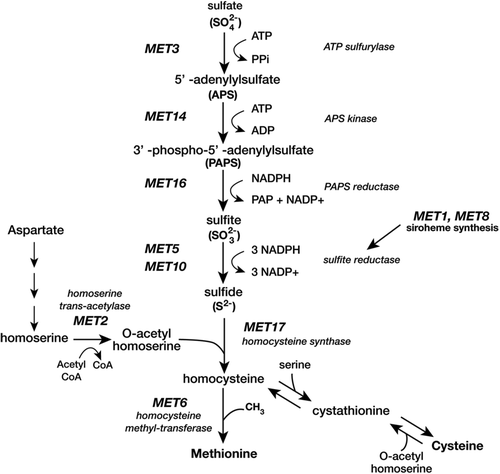
FIGURE 2. Methionine synthesis in S. cerevisiae. The MET genes studied in the class project encode enzymes that catalyze various reactions involved in methionine synthesis (Thomas and Surdin-Kerjan, 1997). Sulfite reductase is a heterotetramer composed of the gene products of the MET5 and MET10 genes. The MET1 and MET8 genes encode enzymes that synthesize siroheme, a cofactor for sulfite reductase.
We selected S. cerevisiae as our genetic model for multiple reasons. It is nonpathogenic, inexpensive, and easily cultured by undergraduate students (Duina et al., 2014; Brownell et al., 2015). Saccharomyces cerevisiae is also a well-studied genetic model. The S. cerevisiae genome was the first eukaryotic genome to be sequenced (Goffeau et al., 1996), and many resources are available to researchers at low cost, including genome-wide collections of strains (Winzeler et al., 1999) and overexpression plasmids (Gelperin et al., 2005). The well-curated Saccharomyces Genome Database (https://yeastgenome.org) is also available for student use.
Although not as widely studied as S. cerevisiae, S. pombe is also a popular model organism (Hoffman et al., 2015) with its own genome database, Pombase (https://pombase.org). Phylogenetic analyses place the two yeast species into different classes that diverged from a common ancestor between several hundred million and a billion years ago (Hedges, 2002; Dujon, 2010). Schizosaccharomyces pombe is considered to be more similar to the common ancestor, while S. cerevisiae has undergone extensive genome diversification, almost certainly facilitated by its domestication by humans (Mortimer, 2000).
Course Structure
BI204 is the introductory laboratory course for biology and biochemistry majors. Students from a variety of other majors also take the course to satisfy premedical requirements. Prerequisites for the course include the molecules and cells introductory lecture class and one semester of general chemistry laboratory. To accommodate the large number of students who need the course, 12 sections of the course are offered each semester. Each section of 15 students is led by a GTA with the assistance of an undergraduate student who performed well in a previous iteration of the course. Sections meet twice per week for 3-hour sessions. Sections are scheduled for four different times during the week. On several occasions during the semester, the three sections scheduled for a particular time come together at the beginning of the session for a short lecture presented by the course director.
The 12 sections of the course work in parallel. Within each section, students work in teams of three. This organization ensures that each student receives hands-on experience with the experimental procedures, because the course experiments are based on groups of three strains, three plasmids, etc. Each team studies one of five different genes involved in methionine synthesis. The five genes selected for study vary from one semester to the next, based on the data of students from previous semesters. GTAs assign students to teams in the second or third class meeting, after the GTAs and undergraduate TAs have had time to observe student group dynamics in the first few laboratory sessions. GTAs are encouraged, but not required, to assign diverse teams based on students’ self-described talents. Students describe their talents on a form modeled after the “guild” system (Wright and Boggs, 2002). In our updated version of the guild system, students are asked to rank the roles they would be best suited to as employees of National Widget Company: the CEO, widget designer, widget marketer, or widget maker.
A few days before the beginning of the semester, GTAs attend a 2-day workshop that includes an overview of the course, an introduction to the pedagogical principles elaborated in scientific teaching (Handelsman et al., 2007), hands-on practice with some course experiments, and a microteaching exercise (Gilreath and Slater, 1974). For the microteaching exercise, GTAs prepare a 5-minute presentation on a topic of their choice in a format of their choice. GTAs present their topics to the course instructors, who provide feedback. Each semester, the teaching staff includes both new and returning GTAs. Returning GTAs are encouraged to share their experiences with new GTAs in common sessions of the workshop, but they are excused from any training sessions they attended in previous semesters. During the semester, all GTAs meet weekly with the course instructors to discuss lab activities for the upcoming week, common problems and misconceptions associated with each lab, assessment processes, and other issues raised by GTAs.
Course Materials
A variety of print and electronic resources have been developed for the course and have been updated on a regular basis in response to GTA and student feedback. 1) A custom laboratory manual provides the theoretical background, experimental protocols, and literature references for course experiments. The manual was developed during the pilot year and has been consistently rated as the most important course material by students. 2) A course site on the university’s learning management system (LMS) provides various supplemental materials for course activities and assignments, including video tutorials, prelab quizzes, assignment rubrics, and links to external tools and databases. The video tutorials were introduced gradually over the first two semesters of the course. During the period described in this report, the LMS changed from Blackboard Vista to Canvas, which provided equivalent services to students. 3) A data-sharing site was developed to archive student data. The data-sharing site is a password-protected Confluence wiki maintained by the university. Students post their data to the site and are able to compare their data with those obtained by students in other sections who are working on the same gene. 4) Students receive a series of primary literature assignments related to an article (Cordente et al., 2009) in which the authors isolate and characterize wine yeast strains that possess mutations in sulfite reductase, a key enzyme in Met synthesis (Figure 2). The assignments were constructed using the CREATE (consider, read, elucidate hypotheses, analyze and interpret the data, and think of the next experiment) approach (Hoskins et al., 2007) and are scheduled to align with experiments in the course research project that employ similar methods or met deletion strains.
Course Activities
Table 1 outlines the class topics and student activities. Our overall goal was to teach core concepts and methods of molecular cell biology to students as they pursued an authentic research question and became more proficient as scientists. The course has a well-defined structure that includes many activities and assessments, a strategy shown to positively affect the performance of students in introductory classes (Freeman et al., 2011). The experimental protocols are designed to be robust, reducing the probability that failures would be due to inherently flawed protocols that can discourage students in UREs (Linn et al., 2015). When an experiment does not “work” as expected, students must decide whether the experiment has yielded a meaningful negative result or whether the result is due to experimental error. To make this judgment, students are encouraged to compare their data with data posted on the data-sharing site by students working on the same genes in other sections. Students may elect to repeat an unsuccessful experiment during open lab week (discussed later in this section).
| Chapter | Topic | Activities |
|---|---|---|
| Module 1. Boot camp | ||
| 1 | Project overview | |
| 2 | Measurement | Students learn about experimental error and how to use micropipettes correctly. |
| 3 | Light microscopy | Students use the microscope to observe and contrast S. pombe and S. cerevisiae cultures in exponential and stationary growth phases. |
| 4 | Yeast techniques | Students learn basic microbiological techniques and the nomenclature conventions for yeast genotypes and phenotypes. |
| 5 | Databases | Students find information on their MET genes in National Center for Biotechnology Information and organism databases. |
| Module 2. Identification of S. cerevisiae met deletion strains | ||
| 6 |
| Students identify mutant strains by their growth properties on various sulfur sources and indicator media. |
| 7 | PCR | Student devise and implement a strategy to distinguish strains by colony PCR. |
| 8 |
| Students analyze their PCR products by agarose gel electrophoresis. |
| 9 |
| Students learn about amino acid chemistries and use the BLAST algorithm to identify conserved regions in proteins encoded by MET genes. |
| Module 3. Characterization of yeast overexpression plasmids | ||
| 10 | Plasmids | Students purify yeast overexpression plasmids. |
| 11 |
| Students select restriction enzymes that will distinguish their plasmids and separate the restriction fragments on agarose gels. |
| Module 4. Functional complementation | ||
| 12 |
| Students transform met deletion strains with overexpression plasmids and use replica plating to determine whether complementation has occurred. |
| Module 5. Molecular analysis of protein expression | ||
| 13 | Protein overexpression | Students prepare protein extracts from transformed cells under both inducing and noninducing conditions. |
| 14 | SDS–PAGE | Students analyze their protein extracts by SDS–PAGE and stain the gels with Coomassie blue. |
| 15 |
| Students analyze expression of overexpressed proteins using antibodies to epitope tags on the Met proteins. The data are combined with SDS–PAGE data in microreport 5. |
The course can be viewed as a series of five modules (Table 1). Numbers on the left of each row in Table 1 refer to chapter numbers in the course manual. The activities outlined in some chapters could be completed in a single class, while others required more than one class to complete. Throughout the semester, students prepare their lab notebooks before each class and take prelab quizzes corresponding to each of the chapters in the manual. In the first “boot camp” module, students learn and practice transferable skills that they will use in later segments of the course or in other lab settings.
In the second module, student teams are given three coded yeast deficiency strains and asked to design a strategy that will identify which of three MET genes is missing in each strain. Students first identify the mutant strains by their ability to grow on various sulfur sources, such as sulfite and cysteine, in place of methionine (O’Connor, 2016). Students then confirm the strain identities using colony PCR. In the third module, students design a restriction mapping strategy to distinguish between three yeast overexpression plasmids. One of the plasmids carries the autologous S. cerevisiae gene to serve as a positive control, and a second plasmid carries the S. pombe orthologue under study. The third plasmid carries an unrelated gene to serve as a negative control. Although instructors know the identities of the strains and plasmids used in these modules, students must select the primer pairs and restriction enzymes that are needed to identify the strains and plasmids.
In the fourth module, students transform the deletion strains with the three plasmids and use replica plating to determine whether S. pombe homologues can substitute for the S. cerevisiae protein that is missing in their deletion strain. Complementation in this experiment provides evidence for functional conservation of enzyme activity between the two divergent yeast species. Failure to complement could indicate lack of functional conservation, but students need to consider alternative explanations. For example, epitope tags encoded in the plasmids might be affecting enzyme function or the protein may not be expressed in the transformed cells. In the fifth module, students prepare protein extracts from the transformed cells and use Western blots to determine whether proteins are correctly expressed in the transformed cells.
Student teams also have the opportunity to design an independent experiment that extends the results obtained in modules 2–4. The experiments are conducted during an open lab week when labs are not held at their normally scheduled times and the lab is open for experimentation during normal business hours. In advance of open lab week, student teams submit an experimental plan that includes a hypothesis, a list of required reagents, and a detailed schedule of activities. Although students are encouraged to propose novel experiments, many teams elect to repeat or improve experiments that generated unanticipated or confusing results earlier in the semester.
Course Assessment
A variety of assessments are used to evaluate student progress toward the course goals of content mastery and the acquisition of research skills. Objective concept tests are used throughout the semester to provide one measure of content mastery. Before each new chapter (Table 1) is introduced in class, students take an online concept quiz consisting of 20 multiple-choice questions. The quizzes are posted 48 hours before class and automatically graded by the course management system.
Students demonstrate both content mastery and research skills in a series of five “microreports” at the end of each course module (Table 1). Each microreport consists of three parts. 1) Student teams prepare a labeled data figure with a descriptive legend and upload the figure to the course data-sharing site. Importantly, these figure legends are strictly descriptive and contain no interpretations of the data. 2) Student teams then present and discuss their data and summary tables in class. 3) Finally, individual students write brief reports that incorporate the group figure and summary tables together with individually written methods and discussion sections. GTAs grade both the oral presentations and written reports using rubrics developed by the course instructors. Students are able to use feedback from GTAs as they prepare their final reports for the semester.
At the end of the semester, students incorporate their accumulated data, including those from the open lab week, into a team poster and a final written report. Teams present their posters during the last class meeting. Three concurrent sections present their data at each poster session, giving students the opportunity to discuss their data and experimental issues with their counterparts from other sections who have worked on the same genes. At the end of the semester, individual students organize their data into a comprehensive report that uses the format of a scientific publication. GTAs grade the final reports and also assign students a participation grade with a rubric that emphasizes student leadership and collaboration skills.
METHODS
In the present study, we used a pre/posttest design to investigate the degree to which enrollment in BI204 was associated with changes in student content knowledge and research methods skills using instruments developed for this project. A content knowledge test and a research methods skills survey were electronically administered during the first and last class meetings. Data were collected over five semesters of implementation (Fall semester of 2012 through Fall semester of 2014), following a pilot year when no data were collected, and were aggregated for a single analysis. To determine whether the course effectively served diverse groups of students, we also examined student growth during the course as a function of students’ personal and academic characteristics (e.g., race/ethnicity, major, gender, and premedical status).
Participants
Each semester, between 170 and 180 students enrolled in the course. Student demographic and academic characteristics were very similar across the five semesters analyzed in this study (Table 2). The total number of students who provided informed consent to participate in this research study was 793. Based on pretest data, slightly more than half the students were female, and white students represented the predominant racial/ethnic group, followed by Asian students and Latino students. Students from URM groups (Black/African American, Hispanic/Latino, and Native American/Alaskan Native) represented 15.7% of the sample. Most of the students in the class were sophomores, followed by juniors, with much lower numbers of seniors and freshmen. Our sample represents students from 26 disciplines, as well as students with multiple majors and students who had not yet declared a major. Despite this academic diversity, students majoring in biology, biochemistry, and psychology comprised most of our sample. The majority of students expressed interest in the premedical program, which is available to students from all majors and requires BI204.
| Percent | Percent | ||
|---|---|---|---|
| Race/ethnicity | Undergraduate level | ||
| Black or African American | 3.7 | Freshman | 1.0 |
| American Indian or Alaskan Native | 0.1 | Sophomore | 73.5 |
| White | 61.2 | Junior | 20.6 |
| Asian | 17.7 | Senior | 4.9 |
| Hispanic or Latino | 11.9 | ||
| Two or more races | 2.9 | Undergraduate major | |
| Nonresident alien | 2.5 | Biology | 55.2 |
| Biochemistry | 17.9 | ||
| Sex | Psychology | 9.0 | |
| Male | 44.7 | Other | 17.9 |
| Female | 55.3 | ||
| Premedical program | 77.9 |
Instrumentation
Data were largely collected via four in-class, electronically administered instruments: a content knowledge pretest; a content knowledge posttest; a pretest survey; and a posttest survey. Of those providing informed consent, the response rates for each of these four instruments were: 92.9%, 96.5%, 89.4%, and 86.1%, respectively.1 Established methods were used to develop and provide validity (Reeves and Marbach-Ad, 2016) and reliability (Kuder and Richardson, 1937; Cronbach, 1951) evidence for the instruments, as detailed more fully in the paragraphs that follow. All instrumentation is available in the Supplementary Material.
Content Knowledge Measurement
The content knowledge test consisted of 19 dichotomously scored multiple-choice items,2 with at least one question matched to each of the course topics (Table 1). For example, there were items pertaining to the concepts of model organisms, yeast mutants, PCR, and Western blots. Explicit alignment of individual test items with course topical domains represents one line of validity evidence (validity evidence based on test content). Factor analysis of the tetrachoric item intercorrelations was conducted at pretest and posttest separately to examine the internal score structure of the test, providing another form of validity evidence (Reeves and Marbach-Ad, 2016). This procedure is used to understand latent dimensions undergirding a larger set of scores (item responses). At pretest, the first factor had an eigenvalue of 6.35 and explained 33.42% of the common item variance, with item factor loadings ranging from 0.37 to 0.70. The factor structure was similar at posttest, with the first factor having an eigenvalue of 6.05 and explaining 31.86% of the variance; item factor loadings ranged from 0.18 to 0.70. These analyses thus suggested that a single common factor (i.e., content knowledge) could explain pretest and posttest content knowledge item scores. This interpretation was supported by Rasch modeling (Rasch, 1960) of posttest item responses from the first semester, which also provided evidence for a single score dimension (unpublished data). Finally, evidence for test validity was obtained from a comparison of posttest content knowledge test scores with course grades, which were shown to be well correlated (r = 0.38, p < 0.001).
KR-20 (Kuder and Richardson, 1937) was used to estimate the reliability of the content knowledge measure, because the items were scored dichotomously. Pretest and posttest reliabilities3 were 0.49 and 0.72, respectively, and the correlation (r) between the pretest and posttest content knowledge measures was 0.23 (p < 0.001). Likewise, Rasch model–based reliability indicators (Rasch, 1960) were also acceptable (unpublished data). As the factor analysis suggested a common factor underlying the content knowledge items, and given acceptable reliability (at posttest particularly), we computed composite pretest and posttest content knowledge measures by taking the means of the items at each time point for each respondent; we also computed content knowledge gain measures by subtracting pretest scores from posttest scores.
Research Methods Skills Measurement
Pretest and posttest surveys consisted of a self-report research methods skills measure. This instrument required each student to indicate his or her level of agreement with 23 statements related to various research methods skills (Table 3). The instrument was originally designed to measure student proficiency in experimental design, collaboration, information literacy (defined as the effective use of the primary scientific literature and databases), written and oral scientific communication, and general technical proficiency. The design of the survey items was informed by prior observational and interview data collected from BI204 students. As with the content knowledge measure, explicit alignment of many statements with the domains of research methods skills targeted by the course provides content-related evidence for the validity of inferences drawn from the instrument (Reeves and Marbach-Ad, 2016). The response format for these items was: “strongly disagree” (coded 1), “disagree” (2), “neither agree nor disagree” (3), “agree” (4), and “strongly agree” (5).
| Itemb | Factorc | Pretest | Posttest | Changed | de |
|---|---|---|---|---|---|
| I feel confident in my ability to construct a testable hypothesis. | ED | 3.85 | 4.15 | 0.30*** | 0.40 |
| I could recognize what a testable hypothesis is in an experimental design. | ED | 4.07 | 4.27 | 0.20*** | 0.28 |
| I could explain what a control is in the context of a scientific experiment. | ED | 4.33 | 4.41 | 0.08* | 0.12 |
| I feel confident that I could design controls for an experiment. | ED | 3.85 | 4.15 | 0.30*** | 0.38 |
| I feel confident in my ability to choose appropriate technology (i.e., methods) to answer a research question. | 3.23 | 3.92 | 0.69*** | 0.84 | |
| I can recognize what goals are realistic for an experiment. | 3.77 | 4.02 | 0.25*** | 0.33 | |
| I feel confident in my ability to use scientific articles as a background for a hypothesis. | 3.67 | 3.87 | 0.20*** | 0.23 | |
| I feel confident in my ability to assemble a bibliography. | 4.09 | 4.06 | −0.02 | -0.03 | |
| I feel confident communicating the results of an experiment to a group of my peers. | OC | 4.01 | 4.16 | 0.16*** | 0.20 |
| I feel confident communicating the results of an experiment to a group of scientists. | OC | 3.19 | 3.58 | 0.39*** | 0.42 |
| I feel confident using technical vocabulary when presenting the results of an experiment. | OC | 3.38 | 3.80 | 0.42*** | 0.49 |
| I feel confident in my ability to write a paper in scientific format. | WC | 3.15 | 3.88 | 0.73*** | 0.80 |
| I feel confident in my ability to write a clear and succinct research paper. | WC | 3.49 | 3.80 | 0.31*** | 0.35 |
| I can recognize when my data have the quality that one expects from published data. | 3.25 | 3.78 | 0.53*** | 0.61 | |
| I feel confident in my ability to produce publication-quality results when I perform an experiment. | 2.92 | 3.50 | 0.58*** | 0.64 | |
| I feel confident in my ability to locate gene-specific information in a scientific database (e.g., National Center for Biotechnology Information). | DB | 2.60 | 3.87 | 1.27*** | 1.35 |
| If I need to locate information about a gene for my experiment, I know where to search for that information. | DB | 2.46 | 4.00 | 1.54*** | 1.70 |
| When working with a group on an experiment, I can effectively divide the tasks between group members. | CO | 4.22 | 4.30 | 0.08* | 0.11 |
| I feel confident in my ability to do research with others. | CO | 4.22 | 4.30 | 0.08* | 0.11 |
| I find it helpful to work with a team when doing research. | CO | 4.14 | 4.22 | 0.08* | 0.09 |
| I feel confident in my ability to work with a team to interpret data from an experiment. | CO | 4.19 | 4.28 | 0.09* | 0.12 |
| I feel confident in my ability to read and analyze scientific papers. | 3.57 | 3.83 | 0.26*** | 0.31 | |
| I feel confident in my ability to understand graphs and tables in scientific papers. | 3.88 | 3.98 | 0.11* | 0.14 |
Common factor analyses (using principal axis factoring) of the research methods skills item scores, separately at pretest and posttest, were used to understand internal structure validity. At pretest and after an oblique rotation, this analysis suggested five substantive factors underlying the item responses: written communication, collaboration, databases, experimental design, and oral communication. Table 3 shows the specific items associated with each factor with loadings greater than 0.50. Eight items, including the primary scientific literature items, had loadings less than 0.5 on all factors. The factors were all positively correlated, meaning that individuals who were “high” on one factor tended to be “high” on the other factors as well (as would be theoretically expected).
A similar exploratory factor analysis of the posttest survey item scores revealed only two factors that were moderately correlated. Substantive analysis revealed that factor 1 concerned research methods skills in general, whereas factor 2 represented collaboration. With the exception of the collaboration skills, these results suggested that individuals tended to respond to the research methods skills in a unitary way at posttest. Because the pretest data suggested five research methods skills dimensions, however, we opted to examine changes in each of the five research methods skills dimensions, as well as overall research methods skills.
Reliability analyses also indicated that the research skills measures were highly reliable at both pretest and posttest. The pretest and posttest reliabilities, as estimated by Cronbach’s alpha (α), for the overall research methods skills measures were 0.92 and 0.97 respectively, and the correlation between the pretest and posttest overall research methods skills measures was 0.18 (p < 0.001). Pretest and posttest reliabilities for item sets constituting each of the five factors (α), were also high and are shown in Table 3. While literature indicates potential limitations of self-reported measures (Falchikov and Boud, 1989; Kruger and Dunning, 1999), the validity and reliability evidence for this specific self-report measure is quite favorable. Therefore, we computed composite pretest and posttest research methods skills measures by taking the mean of the respective items at each time point, and subsequently subtracted pretest scores from posttest scores to yield gain scores for use as dependent variables in the analysis.
The pretest and posttest surveys also gathered student demographic and academic data, including gender, graduation year, major, and enrollment in the premedical program. Race/ethnicity data were obtained from the institutional data warehouse. These data were used in regression analyses designed to uncover differential outcomes for various student populations. Missing data percentages for the content knowledge pretest, content knowledge posttest, research methods skills pretest, and research methods skills posttest scores were as follows: 7.1, 3.5, 10.7, and 13.9, respectively. Given the relatively small amounts of missing data for these variables, the centrality of our research question concerning changes in these variables, and our focus on mean differences, we used mean imputation to handle missing data for these four variables. This treatment does not affect mean values, but it does slightly attenuate the item variance estimates. For the ancillary variables of race/ethnicity, sex, major, and pre–medical student status, missing data ranged from 0.6% to 10.8%. We did not impute data for these variables, but we did use listwise deletion to exclude individuals for whom we did not have complete data (∼19% of respondents) from the regression analyses.
Analytic Approach
We used inferential statistical analyses to examine the degree to which enrollment in BI204 was associated with changes in student content knowledge and research methods skills. We conducted dependent-samples t tests to test whether posttest means were statistically higher than pretest means for content knowledge and research skills. Separate t tests were used to analyze changes in content knowledge, overall research methods skills, and each of the five research methods skills dimensions.
Ordinary least-squares (OLS) regression analyses were also used to examine whether growth in content knowledge and research methods skills varied as a function of student characteristics, such as race/ethnicity, major, sex, and premedical status. In doing so, we effectively investigated whether the course effectively served diverse groups of students. For the regression analyses, gain scores were used as the dependent variable and race/ethnicity, major, sex, and premedical status were used as independent variables. One OLS regression was used to analyze overall gains in research methods skills, and separate OLS regressions were used to analyze each research methods skills dimension (e.g., experimental design, oral communication). Regression analyses of changes in content knowledge were more complicated because of unexpected section-to-section variation4 in the dependent variable, content knowledge gains. An unconditional multilevel model showed significant variance at both level 1 ( , Wald’s Z = 17.12, p < 0.001) and level 2 (
, Wald’s Z = 17.12, p < 0.001) and level 2 ( , Wald’s Z = 3.99, p < 0.001), with an unconditional intraclass correlation (
, Wald’s Z = 3.99, p < 0.001), with an unconditional intraclass correlation ( ) of 0.21. Consequently, this regression analysis was conducted within a multilevel framework (students at level 1 and course sections at level 2; Raudenbush and Bryk, 2002). In the multilevel model, only the intercept parameter was permitted to vary randomly across clusters.
) of 0.21. Consequently, this regression analysis was conducted within a multilevel framework (students at level 1 and course sections at level 2; Raudenbush and Bryk, 2002). In the multilevel model, only the intercept parameter was permitted to vary randomly across clusters.
For the regression analyses all categorical independent variables were dummy coded (e.g., male = 0, female = 1; and not a premedical student = 0, premedical student = 1). Three dummy variables were created to model race/ethnicity: a dummy representing URM students (i.e., Black or African American, American Indian or Alaskan Native, and Hispanic or Latino); a dummy representing Asian students; and a dummy representing “other” students (nonresident aliens and students who were two or more races). White students were the reference group. The major dummy variables were biology, biochemistry, and psychology, with all other majors as the reference category. Also, the dependent variable in each regression analysis was standardized (M = 0, SD = 1) so that regression coefficients for the dummy variables could be interpreted in terms of differences in SD units. Neither bivariate correlations nor collinearity indices (here unreported but available from the authors upon request) suggested multicollinearity problems.
RESULTS
Concept tests and surveys from five semesters were used to elicit evidence of changes in student content knowledge and research methods skills in BI204. Demographic information was then used to determine whether particular groups of students demonstrated differential growth in content knowledge and/or research skills.
Changes in Content Knowledge and Research Methods Skills
Table 3 shows the mean pretest and posttest scores for students’ self-reported research methods skills and changes over the semester. Statistically significant increases were observed for all but one of the 23 research methods skills. Changes were observed in each of the five research methods skills dimensions (discussed earlier) as well as items that did not load highly on any of those five dimensions. As shown by the Cohen’s d values in Table 3, the magnitudes of the changes varied widely across items, suggesting that improvement in some specific skills was greater than for other skills.
Table 4 shows the overall changes in content knowledge and research skills across all semesters, together with research methods skills grouped into the five dimensions identified by the factor analysis. The significance of the gains in each category was tested using paired t tests. Students exhibited statistically higher content knowledge at posttest than pretest, and the magnitude of the gain was very large, with d = 1.85. Students also reported statistically significant overall growth in research methods skills from the beginning of the semester to the end. The magnitude of the reported overall research methods skills change was less than that observed for content knowledge, but was still large, with d = 0.65. Mean changes in each of the five dimensions of research methods skills were also statistically significant. The largest gains were seen for database skills (d = 1.60). Large gains were also observed for written scientific communication skills (d = 0.62). The smallest gains were shown for collaboration (d = 0.12). Interestingly, at the beginning of the semester, students had expressed highest levels of confidence in collaboration and lowest levels of confidence in database skills. At the end of the semester, student confidence in all skills scales was much more similar than at the beginning of the semester.
| Pre | Post | Change | |||||
|---|---|---|---|---|---|---|---|
| Measure | M | SD | M | SD | M | SD | da |
| Content knowledgeb | 0.41 | 0.14 | 0.70 | 0.17 | 0.29*** | 0.19 | 1.85 |
| Research methods skillsc | |||||||
| Overall | 3.63 | 0.48 | 4.01 | 0.65 | 0.37*** | 0.74 | 0.65 |
| Written communication | 3.32 | 0.85 | 3.84 | 0.83 | 0.52*** | 1.11 | 0.62 |
| Collaboration | 4.19 | 0.58 | 4.28 | 0.77 | 0.08** | 0.87 | 0.12 |
| Databases | 2.53 | 0.90 | 3.94 | 0.84 | 1.40*** | 1.17 | 1.60 |
| Experimental design | 4.02 | 0.53 | 4.24 | 0.72 | 0.22*** | 0.82 | 0.35 |
| Oral communication | 3.53 | 0.75 | 3.85 | 0.75 | 0.32*** | 0.96 | 0.43 |
Differential Gains
Regression analysis was used to analyze potential relationships between gains and student characteristics. Table 5 shows regression results for the relationships between gains in content knowledge and research methods skills and student characteristics. For content knowledge gains, it was necessary to use multilevel modeling, because content knowledge gains varied significantly by course section. The fitted model revealed statistically higher gains for URM students during the semester, but no other group differences. The beta coefficient of 0.27 suggests that URM students grew about a quarter of an SD more in their content knowledge compared with white students, who were used as the reference group.
| Research methods skills (N = 655) | |||||||
|---|---|---|---|---|---|---|---|
| Content knowledge (N = 646) | Overall | WC | CO | DB | OC | ED | |
| Regressor | βstd. (SE) | βstd. (SE) | βstd. (SE) | βstd. (SE) | βstd. (SE) | βstd. (SE) | βstd. (SE) |
| URM race/ethnicity | 0.27 (0.10)* | 0.01 (0.12) | 0.04 (0.12) | −0.07 (0.12) | −0.21 (0.12) | 0.10 (0.12) | 0.12 (0.12) |
| Asian race/ethnicity | 0.11 (0.10) | −0.02 (0.11) | 0.00 (0.11) | 0.13 (0.10) | −0.21 (0.11) | 0.06 (0.11) | 0.00 (0.11) |
| Other race/ethnicity | −0.32 (0.17) | −0.03 (0.18) | 0.03 (0.19) | −0.10 (0.18) | −0.20 (0.19) | 0.06 (0.19) | 0.16 (0.19) |
| Biology major | −0.08 (0.10) | −0.12 (0.11) | −0.01 (0.11) | −0.27 (0.11)* | −0.06 (0.11) | 0.00 (0.11) | −0.23 (0.11)* |
| Biochemistry major | −0.01 (0.12) | −0.24 (0.13) | −0.12 (0.14) | −0.43 (0.13)** | 0.01 (0.13) | −0.14 (0.14) | −0.41 (0.14)** |
| Psychology major | 0.13 (0.16) | −0.08 (0.17) | 0.02 (0.17) | −0.22 (0.17) | −0.06 (0.17) | 0.00 (0.17) | −0.24 (0.17) |
| Female | −0.08 (0.07) | 0.13 (0.08) | 0.09 (0.08) | 0.12 (0.08) | 0.09 (0.08) | 0.13 (0.08) | 0.14 (0.08) |
| Premedical | −0.07 (0.09) | −0.01 (0.10) | 0.04 (0.10) | −0.01 (0.10) | −0.01 (0.10) | −0.10 (0.10) | −0.03 (0.10) |
| Model | χ2(8) = 4.47, p = 0.81 | F(8,646) = 0.85, p = 0.56 | F(8,646) = 0.38, p = 0.93 | F(8,646) = 1.97, p < 0.05* | F(8,646) = 0.93, p = 0.49 | F(8,646) = 0.84, p = 0.57 | F(8,646) = 1.86, p = 0.06 |
| R2 | 0.00 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.02 |
Table 5 also shows the results from the OLS regression analyses used to elicit potential relationships between student characteristics and gains in both overall and each of the five dimensions of research methods skills. Unlike content knowledge gains, self-reported research methods skills gains did not systematically vary by course section, so multilevel modeling was not necessary. The regression model for reported gains in overall research methods skills showed no significant association with students’ characteristics. Likewise, gains in written communication, oral communication, and database use were not associated with particular student characteristics. Notably, students who were not majoring in biology or biochemistry showed significantly stronger growth in collaboration skills and marginally stronger growth in experimental design skills.
DISCUSSION
Undergraduate research experiences (UREs) have been advanced as a mechanism by which to improve undergraduate STEM education (e.g., Eagan et al., 2013). However, resource constraints often impede institutions of higher education from providing traditionally conceived UREs to large numbers of their students, and most particularly, to introductory students. CUREs offer an alternative mechanism for expanding access to authentic research experiences to larger and more diverse groups of students (Auchincloss et al., 2014; Bangera and Brownell, 2014; Spell et al., 2014; Brownell and Kloser, 2015). This study describes a large introductory CURE that incorporates a research project in functional genomics together with a multidimensional evaluation of the changes observed in student knowledge and research methods skills over five semesters.
The evidence collected from the large, multisemester sample reported here suggests that participation in the course is associated with sizable growth in students’ content knowledge and research methods skills (Table 4). Gains were particularly robust for student content knowledge, which was assessed with a concept test aligned with the custom lab manual for the course. The large magnitude and specificity of the content knowledge gains make it unlikely that they are simply due to other factors such as maturation or concurrent non-BI 204 learning experiences. The magnitude of student gains in content knowledge was similar for all student populations, with the notable exception of URM students (i.e., Black/African American, Hispanic/Latino, and American Indian/Alaskan Native students), who demonstrated greater growth in content knowledge than white students. While URM students demonstrated less content knowledge at pretest than did their white counterparts, this finding is consistent with those of others who have documented the role that research experiences and small class communities can play in URM performance and retention (Jones et al., 2010; Hernandez et al., 2013).
These gains in content knowledge were unexpectedly complex, however. Multilevel modeling revealed systematic variation in content knowledge gains across class sections. Some of these differences may be attributable to the GTAs who taught the individual sections. Over the five semesters of this study, the sections were taught by 30 different GTAs, each of whom taught the course anywhere from one to four semesters. The GTAs were quite diverse with respect to their own educational backgrounds and research interests. Another factor that may have contributed to the differences between sections is the nonrandom enrollment of students in the various sections, which is governed by a seniority system.
Across five semesters of implementation, students also demonstrated gains in all research methods skills (Table 3), with the single exception of preparing bibliographies. Roughly two-thirds of the research methods skills on the student survey mapped onto one of five factors. The largest gains were consistently observed for database-search skills. BI204 probably provided most students with their first opportunity to interact with online databases for genetic information, as evidenced by the lowest confidence ratings at pretest. The next highest gains were associated with written communication, followed by oral communication. This may reflect the course structure, which is organized around five data microreports that incorporate several levels of feedback on student work. At each data reporting point, teams give a group oral presentation to their sections. Individual students may choose to incorporate the class feedback as they prepare individually written reports, which are graded by the GTAs. At the end of the semester, students compile their data into a group poster and individually written final reports in the format of a scientific publication. Student feedback has been consistently positive about the value of the microreports.
Students also reported increases in their experimental design skills over the course of the semester. Although students largely used established protocols, laboratory activities were designed to incorporate elements of inquiry, and students had the opportunity to design an original experiment during open lab week. The smallest, but nonetheless significant, gains were associated with student collaboration skills. This may reflect the fact that students began the semester with high levels of confidence with respect to collaboration.
Students reported some of the highest gains for skills that did not map onto one of the five common factors (Table 3) revealed in our analysis. For example, skills relating to choosing appropriate technology, recognizing realistic goals for experiments, and producing publication-quality data were associated with significant gains. Other gains that did not map to common factors were related to use of the primary literature. For our literature assignment, we adapted the CREATE method (Hoskins et al., 2007) to a research article on met mutations that reduce the sulfide produced by wine yeast (Cordente et al., 2009). The article was chosen because it involves the Met biosynthetic pathway and uses some of the same strains used in the class research project.
The gains that we observed in research methods skills were largely similar across student groups (Table 5), with two exceptions. The analysis indicated that students who were majoring in biology and biochemistry grew slightly less than nonmajors in skills related to collaboration and experimental design. This may reflect the fact that biology and biochemistry majors are likely to have taken more laboratory courses before or concurrent with BI204. Unlike content knowledge, gains in research methods skills were statistically indistinguishable for students of different racial/ethnic groups.
The course that we have described here incorporates many characteristics of CUREs (Auchincloss et al., 2014; Corwin et al., 2015b). In researching the functional conservation of Met biosynthetic enzymes between evolutionarily divergent yeast species, students are collectively pursuing an authentic research question of broad relevance. Although Met synthesis has been well studied in S. cerevisiae (Thomas and Surdin-Kerjan, 1997), functional information is still not available for most of the S. pombe homologues. BI204 students have successfully demonstrated homology for most of the S. pombe genes shown in Figure 2. These data are now being finalized for a scientific publication by BI204 alumni enrolled in an advanced lab class.
The data in Tables 3 and 4 provide evidence for student growth in both collaboration and scientific practices. Iteration is the CURE element that was most difficult to incorporate into our course design because of constraints related to the large number of students, content learning goals, and scheduling considerations. Recognizing the importance of iteration to mastery, our experiments were designed so that students would use the same techniques, for example, spot plating and agarose gels, multiple times during the semester. With the exception of open lab week, students are not able to repeat individual experiments. We therefore use our data-sharing site to provide a kind of virtual iteration. Students are able to view the experimental data obtained by other students pursuing the same question in different sections.
LIMITATIONS AND FUTURE DIRECTIONS
The present study has a number of limitations common to biology education research. Our analyses have documented significant gains in content knowledge and self-reported research skills for students in BI204, but there is no control course(s) for comparison. Our statistical models have shown that, on average, students grew by a large amount, but our analysis also revealed significant amounts of unexplained variation both between and within sections of the course. It will be important to identify sources of systematic variation in the context of other student characteristics or external factors, such as classroom composition or GTAs. Finally, our investigation has measured changes in student content knowledge and research skills over the course of a single semester. It would be interesting to determine whether participation in BI204 has longer-term outcomes such as undergraduate retention and interest in graduate school. These limitations notwithstanding, the present evaluation was rigorous in that it employed multiple measures, and data were collected from a large, multisemester sample. It seems unlikely that the observed gains, which were especially large for content knowledge, can be explained by other factors such as maturation.
We hope that educators at other institutions will be interested in adapting this project to their own institutions. The project was deliberately designed to be inexpensive to implement and easily adapted to other institutional settings. We have shown here that the project is effective as a stand-alone, 3-credit CURE, but we recognize that instructors may also be interested in using portions of the course in existing lab classes. Many of our course materials are available at the BI204 website (https://capricorn.bc.edu/bi204). We have already shared these and other course resources with colleagues at several other institutions who have successfully incorporated parts of the S. pombe project into existing 1-credit lab classes. At Hampden Sydney College, for example, students in a sophomore-level genetics lab demonstrated increases in research skills similar to those described here (Wolyniak et al., 2013). Some of the course topics, such as the genetic analysis of sulfur sources in Chapter 6 (Table 1), can also be used as an individual lesson (O’Connor, 2016).
The project framework described here can be adapted to a variety of other genetic systems and research questions (Figure 1), informed by the expertise and research interests of course instructors. We have found S. cerevisiae to be a particularly amenable test organism, but similar experiments should be possible using bacteria or simple eukaryotes like Caenorhabditis elegans as test organisms. The project framework is a dynamic one that will naturally require modifications as research questions are answered. At Boston College, for example, students have demonstrated functional conservation of almost all the MET genes in the pathway shown in Figure 2 between S. cerevisiae and S. pombe. We have therefore modified the course project design to use the filamentous yeast Neurospora crassa as the source of potential MET gene homologues. The N. crassa genome is approximately twice the size of the S. cerevisiae genome, with paralogues for several MET genes. We hope that others will use the framework that we describe here to develop other research projects that engage students in genomics research.
FOOTNOTES
1Three participants with implausibly short completion times for the content knowledge test were excluded from analyses related to that variable.
2There was initially a 20th content knowledge item concerning methionine, which was subsequently replaced (in Fall 2014) with an item related to sulfur, because an item analysis indicated problems with its properties; due to comparability issues, both of these items were excluded for the content knowledge analyses conducted for this paper.
3The KR-20 calculation is based on a ratio of variances: the sum of the item variances over the total score variance. When a pretest of content knowledge is relatively hard, as is the case here, the item variances will be small, because most people get the items wrong—hence, the KR-20 is weak. A measure of effective change is demonstrated when the KR-20 improves at posttest, because now there is greater variation on the items.
4Preliminary analyses showed that there was no empirical clustering in the research methods gain variable (i.e., research methods gains did not vary systematically by course section), and thus multilevel modeling was not necessary.
ACKNOWLEDGMENTS
We thank all students who participated in the project and the BI204 staff for their help in administering the assessment instruments. We thank Dr. Katie Trong for her help in developing the student research skills instrument. This project was supported by National Science Foundation grant 1140428.



