Evolving Strategies for the Incorporation of Bioinformatics Within the Undergraduate Cell Biology Curriculum
Abstract
Recent advances in genomics and structural biology have resulted in an unprecedented increase in biological data available from Internet-accessible databases. In order to help students effectively use this vast repository of information, undergraduate biology students at Drake University were introduced to bioinformatics software and databases in three courses, beginning with an introductory course in cell biology. The exercises and projects that were used to help students develop literacy in bioinformatics are described. In a recently offered course in bioinformatics, students developed their own simple sequence analysis tool using the Perl programming language. These experiences are described from the point of view of the instructor as well as the students. A preliminary assessment has been made of the degree to which students had developed a working knowledge of bioinformatics concepts and methods. Finally, some conclusions have been drawn from these courses that may be helpful to instructors wishing to introduce bioinformatics within the undergraduate biology curriculum.
INTRODUCTION
The Importance of Bioinformatics within the Undergraduate Biology Curriculum
The publication of the first draft of the human genomic DNA sequence in 2001 (Lander et al., 2001; Venter et al., 2001) heralded a new era in biology. The life sciences continue to be transformed by the rapid accumulation of a rich array of data of diverse types. In order to access and exploit this information, biologists have become increasingly dependent on computational approaches to access, annotate, and analyze these data sets—that is, the goal of bioinformatics. The importance of bioinformatics to the future of biology is reflected by its inclusion among the new topics covered in recent molecular and cellular biology textbooks (e.g., Lodish et al., 2000, Sect. 7–4). The challenge for biology faculty is to find ways to introduce undergraduate biology students to the basic concepts, tools, and databases in bioinformatics. This objective has been the focus on several recent symposia and workshops (e.g., genomics.wheatoncollege.edu/workshop/).
Several strategies have been employed to incorporate bioinformatics within the undergraduate cell biology curriculum at Drake University. This paper describes the context of these efforts, the specific objectives, and specific examples of activities. Finally, a critical assessment of these strategies is offered, as well as some practical suggestions based on the course experiences to date.
The Context of These Efforts
Drake University is a midsized midwestern university with an enrollment of about 5,000 students. The primary author is an associate professor in the Department of Biology and is responsible for teaching courses in cell biology and molecular biology (along with associated laboratories). These two courses attract students majoring in biology, psychology, and chemistry, as well as those enrolled in interdisciplinary programs in biochemistry—cell and molecular biology, pharmaceutical sciences, and neurosciences. Each of these courses has an enrollment of 40–60 undergraduate students at present, and about half of these students take the associated laboratories. Of these students, the majority (up to 70%) are sophomores. For many of these students, this is the first biology course that they take following an inquiry-based introductory biology sequence in their freshman year. Most of these students have completed course work in general chemistry and are concurrently enrolled in organic chemistry.
In addition, the author has developed elective courses in structural biology and computational biology, and the latter course is described in detail in this paper. A strategy to incorporate bioinformatics throughout the undergraduate biology curriculum has been realized through exercises and projects carried out by students in both introductory and advanced courses.
Fundamental Objectives in Bioinformatics Education
Bioinformatics as a discipline has seen rapid growth in the last several years. Programs in bioinformatics are now being offered at both the graduate and the undergraduate level, at both small colleges and research universities ( biotech.icmb.utexas.edu/pages/bioinform/biprograms_us.html). It is still in its infancy as a discipline, and many fundamental problems remain to be solved by students now entering these programs ( genes.mit.edu/burgelab/topten.htm). A student of bioinformatics will need to develop proficiency in both biology and computer science in order to be competitive in today's job market.
It should be noted that this is not the career goal of most of our students. The majority of our students are interested in pursuing careers in biology or the health sciences. For these students, the objective is not to prepare them for a career in bioinformatics, but to give them literacy in the basic methods and applications of bioinformatics. The goal is to teach these students to be effective users of the vast repository information deposited in biological databases. It is likely that these students will access this information in the workplace or as private citizens.
A subset of these students will go on to pursue careers in molecular and cellular biology in academia or industry. It is expected that the majority of these students will apply these skills within the context of traditional“ wet lab” research. For these students, the goal was for them to acquire the ability to work from DNA sequence to protein structure and function—and back again. Many of us developed this capability during the course of our research training in a piecemeal fashion. It would be advantageous for these students to enter graduate programs or industry with an effective working knowledge of bioinformatics. It is also essential that these students gain a deeper and more critical understanding of bioinformatics methods and their applications, so that they have an appreciation of the strengths and limitations of the tools they use in their work. Therefore it is important that these students develop some understanding of how bioinformatics tools are developed and deployed over the Web.
Developing a Basic Toolbox for Undergraduates Studying Bioinformatics
An instructor's ability to provide students with practical experiences in bioinformatics has been dependent on the technology readily available to students. Dramatic changes in computer technology, and the proportion of students with ready access to it, have taken place since the author began teaching in 1995. The rapid pace of these changes has been as exciting as it has been challenging, and it is clearly only the beginning of the transformation of society in general, and education in particular. Given the dynamic nature of these advances, and the diversity of ways in which students have access to personal computing and the Internet, how is it possible to provide students training in bioinformatics? How does an instructor deal with the different computing environments used by individual students?
The first solution to this problem was to use campus computer laboratories to provide a common computing environment for all of the students. However, this solution proved to be problematic for a number of reasons. Students found it difficult to gain access to these facilities outside of class time, or if they had access, a variety of software or hardware problems interfered with the conduct of their work. In order to address this problem a strategy has been evolved where demonstrations in the classroom or computer laboratory are used to familiarize students with programs that they will subsequently download and run on their own computers.
Through trial and error several cross-platform software packages have been identified that provide students with a “toolbox” for bioinformatics. Students are asked to download the software appropriate to their personal computer's operating system at the beginning of the semester. This bioinformatics toolbox contains some basic utilities: software for editing text and graphics, a molecular graphics viewer, and so on. These tools, and the URL of their sources, are listed in Table 1.
These tools have been chosen because versions exist for computers running Windows, Macintosh, or UNIX-based operating systems such as Linux. In addition, for each category, at least one example is freeware, obviating the need for students to buy additional software for these courses. This does not mean that students cannot use commercial software for these tasks; it just means that they can carry out the assigned course work without the purchase of additional software. And in some cases the freeware solution is superior—using Microsoft Word for the editing of sequences can lead to problems if the files are not deliberately saved in a simple ASCII text format.
This situation has continued to improve, and within the last several years a large number of bioinformatics-oriented sites have appeared on the Web. The key sites used in our courses are listed in Table 2. The easy access to these tools has made the teaching of bioinformatics feasible on campuses lacking the resources to license the proprietary software commonly found in laboratories at research universities and in industry. The ability of undergraduate students to learn how to use the basic tools of bioinformatics will only help them appreciate the convenience of commercial bioinformatics tools that they will encounter as they progress in their careers.
| General databases and tools for bioinformatics studies |
| National Center for Biotechnology Information |
| http://www.ncbi.nlm.nih.gov/ |
| • BLAST |
| http://www.ncbi.nlm.nih.gov/BLAST/ |
| • PubMed |
| http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=PubMed |
| • Online Mendelian Inheritance in Man (OMIM) |
| http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db OMIM |
| • NCBI Conserved Domain Search |
| http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| • CDART: Conserved Domain Architecture Retrieval Tool |
| http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd=rps |
| Protein Data Bank |
| http://www.rcsb.org/pdb/ |
| Access points for integrated suites of sequence analysis tools |
| BCM Search Launcher |
| http://searchlauncher.bcm.tmc.edu/ |
| European Bioinformatics Institute |
| http://www.ebi.ac.uk/index.html |
| •ClustalWa |
| http://www.ebi.ac.uk/clustalw/ |
| Biology Workbench |
| http://workbench.sdsc.edu/ |
| Some resources for human genomics |
| The Human Genome (NCBI) |
| http://www.ncbi.nlm.nih.gov/genome/guide/human/ |
| Human Genome Browser Gateway (UCSC) |
| http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg10 |
| Example of a specialized structure prediction tool |
| COILS Server |
| http://www.ch.embnet.org/software/COILS_form.html |
EVOLUTION OF THESE STRATEGIES
The goal of introducing bioinformatics in the undergraduate biology curriculum was implemented in three stages from 1995 to 2001. The first exercises were introduced in the cell biology laboratory in the fall of 1995. Additional exercises were developed during the second offering of a course in structural biology in 1999. Finally, a dedicated course in bioinformatics was offered for the first time in the spring of 2001, at the same time that the first draft of the human genome sequencing project was published in Science and Nature. These efforts were not independent of one another, and what has been learned in one course has strongly influenced the ongoing development of the other courses. It has also led to the development of different strategies to accomplish the goals described above. Specific examples of how these courses were implemented and refined follow.
Introductory Cell Biology Course
The author was still in the process of acquiring the equipment needed to set up the teaching laboratory during his first semester as an assistant professor. In addition to the wet lab experiments carried out during this semester, several laboratories were conducted using pencil and paper in the laboratory or in 1-h sessions at one of the computer laboratories on campus. It was not reasonable at this time to assume that most of the off-campus students had ready access to the Internet, and at this time there was considerable competition for access to the limited number of Internet-linked computers in these laboratories. The simple exercises described in Table 3 can be incorporated within the classroom as an introduction to bioinformatics, without a requirement for extended access by students to an Internet-capable computer.
| 1. Pencil-and-paper exercise that illustrates the central dogma of molecular biology using the human H-Ras gene |
| In this exercise students were given a two-page handout containing the coding strand DNA sequence containing the human H-Ras gene (GenBank accession number J00277; also accessible as a Adobe Acrobat document, human_hras.pdf), the coordinates of the coding segments, and a copy of the standard genetic code dictionary. Students were given a practical demonstration of the relationship between DNA sequence and protein structure, by carrying out the conceptual translation of this gene. This assignment was completed as a group exercise within about 60 min. The students were introduced to a variety of important concepts such as the central dogma, eukaryotic gene organization (for a split gene), the relationship between the sequence of the coding strand of DNA and the derived mRNA molecule, and the nature of the genetic code. Students come away from this exercise with a better understanding of how proteins are encoded by the sequence of bases in the DNA molecule. |
| 2. Experience with the use of BLAST as a gene identification tool |
| Another early example of an exercise had the students identify the source of an unknown protein or a DNA sequence when they were supplied with either a protein or a DNA sequence. As an example, students were given the amino acid sequence of another member of the Ras family of GTPases. Alternatively, students determined the DNA sequence of the gel in Figure 10.9 of Alberts et al., Essential Cell Biology, and used the derived DNA sequence in a BLAST search. In this exercise students learned how a relatively short DNA or protein sequence could be used to identify and retrieve a database entry containing the entire DNA or protein sequence. |
| 3. Use of RasMol to visualize the location of affected amino acids in oncogenic forms of Ras |
| Students were introduced to the RasMol molecular graphics program in the computer laboratory. Students were instructed as to how to load the atomic coordinates for the Ras protein (1p21.pdb), which had been downloaded from the Protein Data Bank ( www.rcsb.org/pdb/). Students were shown how to alter the display of the protein molecule to highlight the portion of the molecule affected by common oncogenic mutations in Ras. In this exercise students were able to see how the location of oncogenic mutations in Ras correlated with the location of the bound guanine nucleotide and key amino acid residues required for the GTPase activity of this protein. |
| Software required: Web Browser, RasMol |
| Useful Web sites: BLAST @ NCBI, RCSB |
The first exercise in Table 3 has given students insight into the nature of gene structure and expression and, as such, illustrates how exposure to bioinformatics can help students better understand fundamental concepts in molecular cell biology encountered in the research literature. Students in the cell biology course were given a one-page handout containing the DNA sequence of a 6.6-kb BamHI fragment from the genome that contains the human H-Ras gene. This piece of DNA transforms NIH 3T3 cells to cancer cells when only a single nucleotide in codon 12 of the Ras gene is changed from G to T, resulting in the substitution of valine for glycine in the encoded protein (Tabin et al., 1982). As they work together in groups to carry out the conceptual translation of the gene from the nucleotide sequence, students occasionally make mistakes, making it necessary for the group to backtrack or even start over. It impresses them to see how precise the cellular machinery must be to faithfully duplicate and express the information in DNA. As they work through this exercise, they also encounter what is often a surprise—an intron that interrupts a codon. Once again, they see how precise the cell must be, in terms of splicing the transcribed RNA. Finally, they see a remarkable aspect of human gene organization: only about one tenth of this genomic fragment encodes the H-Ras protein.
It is now possible to assume that nearly every student has ready access to a personal computer and the Internet (both on and off campus). This situation has been exploited in that additional Web-based exercises have been developed to acquaint students with the diversity of information available on-line as a result of the genomics revolution in biology. The development of these new exercises has been closely tied to activities in the introductory cell biology course, in particular, the course's required discussion section. In this discussion section students read seven papers that describe key aspects of the cell biology of the Ras gene and its role in cancer.
In the last 2 years, these Web-based exercises have been a required assignment in the cell biology course. These exercises (with hyperlinks) can be accessed at www.cellbio.drake.edu/ASCB/present.html. These exercises focus on various aspects of the Ras gene and protein, giving students specific examples of how these tools can be applied to give insight into the function of a protein that has obvious relevance to human medicine.
An Elective Course in Structural Biology
Students responded very favorably to these early exercises, especially those involving molecular visualizations through the use of the RasMol program. To cultivate this interest, a course in structural biology was subsequently developed to give interested students an opportunity to further explore the relationship between the three-dimensional structure and the biological function of cellular macromolecules. In the fall of 1997 this course in structural was offered for the first time, and it has been offered three more times in subsequent years. In its first manifestation the course work focused on the use of molecular graphics and model building activities as a way for students to gain insight into the function of protein molecules. In 1997 student use of the Web was largely confined to finding and downloading relevant PDB files. The goals, organization, and activities of this course in structural biology will be described in detail in a separate paper (in preparation).
By the time the course was offered a second time in the spring of 1999, the technological context had changed such that a more comprehensive approach to the study of protein structure, function, and evolution could be attempted. In this second iteration, much more emphasis was placed on the use of Internet-based tools to analyze and relate a protein's amino acid sequence to its structure and function. The course organization also changed at this time to represent the author's current strategy in teaching bioinformatics and structural biology. First, students are introduced to tools and databases via simple exercises and assignments during the beginning of the semester. Students then apply these methods to a major individual or group research project during the latter portion of the semester. The students' research culminates in class presentations and final, formal written reports.
In the last several years, this course has emphasized the development of the bioinformatics skills most relevant to structural biology. In addition to the visualization activities inherent in this course, students now carry out a variety of activities that require the development of basic bioinformatics skills. Students use on-line tools to compare the sequences and three-dimensional structures of homologous proteins. Examples of training exercises and research projects used in this course, which are relevant to the development of bioinformatics skills, are listed in Table 4.
| Training exercises |
| Before the following exercises, students were given reading assignments from reviews relevant to the particular class of structure or structural element being examined. |
|
| Research projects |
|
| Software required: Web Browser, RasMol, TreeView |
| Useful Web sites: BLAST @ NCBI, Clustal W @ EBI, RCSB, COILS, VAST |
Structural bioinformatics will certainly develop as an important subdiscipline of bioinformatics, especially as structural genomics efforts come to fruition (Burley et al., 1999). Within the last year an excellent overview of this important aspect of bioinformatics has been published (Bourne and Weissig, 2003).
An Elective Course in Bioinformatics
In the spring of 2001, an introductory course in bioinformatics, entitled“ Computational Biology,” was offered for the first time at Drake University. Although the terms bioinformatics and computational biology are sometimes used as synonyms, the usage has evolved such that the latter term would seem to be more appropriate for describing the use of computational methods in the modeling and simulation of biological systems ( grants1.nih.gov/grants/bistic/CompuBioDef.pdf; Noble, 2002). In contrast to the structural biology course, the introductory bioinformatics course emphasized the process of deducing protein structure and function (when possible) from the genomic DNA sequence of an organism. The emphasis in this course was on sequence analysis of the sequences of DNA and protein molecules.
The course organization was slightly different from that used in the structural biology course. First, a series of training exercises was carried out, but this time in a computer laboratory in the presence of the instructor. The objectives of these initial computer laboratory sessions were for the students to learn what bioinformatics tools were available and to understand what they can be used to do. These activities were conducted in a computer laboratory to see if the instructor could help students address some of the common questions arising from the use of these software tools. Some examples of these training exercises are listed in Table 5. Typically there would be a follow up assignment that would be carried out on the students' personal computers before the next class meeting.
| Training exercises |
|
| Group research project |
| Sequence and structural analysis of a large, multidomain protein (human ryanodine receptor genes/proteins). See the text for a description of this exercise. |
| Group programming project |
| Development of a Web-based Perl program that determines the amino acid composition of any protein sequence in FASTA format. See the text for a description of this exercise. |
| Software required: Web Browser, RasMol, TreeView |
| Useful Web sites: Most of the sites listed in Table 2 |
In addition to these training sessions, students were also given reading assignments in the course textbook (Baxvanis and Ouellette, 2001) and from recent reviews (e.g., Vukmirovic and Tilghman, 2000). The students were also asked to read through the on-line documentation for a program such as BLAST ( www.ncbi.nlm.nih.gov/Education/BLASTinfo/guide.html) or to work through on-line tutorial for a program such as Cn3D ( www.ncbi.nlm.nih.gov/Structure/CN3D/cn3dtut.shtml). Although students were usually content to accept the default parameters for a BLAST search, it is important that they understand some of the underlying theory behind the program's function, that they understand how to correctly and critically interpret the results, and that they understand when and why it may be necessary to change the default parameters. It should be understood that it was not intended that the students would end up with a graduate-level understanding of these programs, but that they would at least start to think critically about the meaning of the results obtained.
One of the issues addressed in this course was why some problems in bioinformatics are difficult to solve. Given the publicity surrounding the publication of the first draft of the human genome sequence in the spring of 2001, one such problem was identified: How many genes are there in the human genome? Why is it that not everyone agrees on the answer to this question? Questions like this are very valuable in that they highlight the limitations of our understanding and in that they serve to present new opportunities for advancement.
Toward the very beginning of the course students carried out an exercise that gave them insight into the nature of this problem. Students were asked to use on-line tools (e.g., www.ncbi.nlm.nih.gov/gorf/gorf.html) to predict the position and extent of open reading frames in two pieces of DNA, one from the human genome containing the H-Ras gene (accession number J00277) and another from E. coli containing the lac operon (accession number J01636). They were asked to compare this information to the known coding sequence coordinates for the encoded gene products. From this exercise the students gain a perspective on how hard it is to identify and define the coding segments for a split eukaryotic gene like human H-Ras without additional information being provided, such as the corresponding cDNA sequence. After this exercise students were asked to look at more sophisticated approaches to eukaryotic gene prediction, such as the GrailEXP program ( compbio.ornl.gov/grailexp/). This type of exercise also demonstrates the differences in gene structure typically seen when eukaryotic and eubacterial genomic DNA sequences are compared.
This exercise also gave students the opportunity to see examples of the type of exceptions to rules that molecular biologists are familiar with but that puzzle the beginning student. One example of this type of anomaly is seen for the lacA gene, where the start codon is UUG instead of the expected AUG (according to the annotation for this gene). Students were also initially puzzled to see the order of the genes in this sequence file (J01636) being apparently backward compared to what they have seen in their textbooks, until they realize that the sequence represents the complement of the coding strand for the genes of the lac operon. This emphasizes to them that the coding strand for a given gene may be the template strand for a neighboring gene. Students may have encountered these points in textbooks or lectures, but real-world examples provide concrete illustrations of these principles.
After carrying out these training exercises in the first third of the semester, students began work on a group research project that applied these tools and concepts. Two groups of about five students worked together to analyze DNA and protein sequence the same type of protein, the ryanodine receptor, an enormous ion channel found in skeletal muscle sarcoplasmic reticulum (Takeshima et al., 1989). Students were asked to take advantage of the data that had recently been submitted to the NCBI sequence database as a result of the human genome project. They were instructed to focus their research on the three members of the ryanodine receptor gene family in humans (RYR1, RYR2, and RYR3).
Their group research project emphasized the integration of information gleaned from various informatics resources: the biomedical literature, the genomic databases, and the structure databases. Students were asked to use the tools encountered in the training exercise, as well as any suitable tools they found on the Internet. Students were asked to divide up the responsibilities for the different types of analyses to be carried out. This instructor met with the students regularly throughout the remainder of the semester to assess their progress and to help them troubleshoot problems they had encountered.
One other question was addressed in the initial training exercises: Are some programs superior to others in terms of the validity of their output? Students were asked to test the success of a variety of programs that predicted the location of transmembrane segments in integral membrane proteins. This experience would have obvious benefit when they analyzed the ryanodine receptor protein sequences. Students were asked to use a variety of programs (e.g., www.ebi.ac.uk/~moeller/transmembrane.html) to predict the transmembrane segments in bacteriorhodopsin, a well-studied membrane protein for which this information was available. By comparing predictions of these different programs with the biochemical and structural studies of this protein, students could see which programs performed better than others for this task.
This naturally leads to the question, What makes some computer programs better than others in accomplishing a specific task? The answer is, of course, the nature of the computer algorithm employed by the program, as well as the underlying assumptions that led to the development of the algorithm. In order to give students a better appreciation of the importance of algorithms in bioinformatics, students worked in two groups to develop a very simple sequence analysis program, which is described in the next section. An additional goal of this particular assignment was that they would develop an appreciation of what happens after they click the “Submit” button on the Web page of a typical sequence analysis program. While it is certainly not necessary that students become computer scientists in order to effectively use bioinformatics programs, an acquaintance with the process of designing, writing, and testing a simple bioinformatics program was intended to demystify the process while providing some understanding of the ways in which even simple computer programs can go astray.
PROGRAMMING A SIMPLE BIOINFORMATICS APPLICATION IN PERL
The most experimental (and perhaps risky) aspect of this bioinformatics course was having the students develop a simple sequence analysis computer program. It was not clear at the beginning how difficult it would be for the students to achieve this goal. Several observations made this author believe that it would be reasonable for them to try to develop this program. First, since most of the enrolled students were biology majors, it seemed reasonable that at least some of them may have taken a college-level programming course as one way to fulfill a mathematics requirement for our major.
A survey was conducted at the very beginning of the course to assess how many of the enrolled students had prior experience with computer programming. In a class of 10 students, 2 had a fair amount of experience with programming, and a few others had some limited experience. Finally, it was the author's earnest belief that all of the students could at least think about the logic of the program, even if they did not have previous experience with encoding this logic into the form of the algorithm used by the program.
The first issue faced in designing this assignment was what would be a suitable task for this program. The task had to be simple since time was limited, but it also needed to accomplish something of use to the students. In the end it was decided that the students would construct a program that calculated the percentage composition of amino acid residues from any protein sequence supplied in the FASTA format ( www.ncbi.nlm.nih.gov/BLAST/fasta.html). The program would simply need to tally the number of each type of amino acid residue present and the total number of residues, use this information to calculate the percentage composition for each residue, and then report the result back to the user. Many programming books start with a programming exercise in which a very simple program generates the text “Hello, world!” This program can be thought of as a kind of “Hello, world!” program for bioinformatics.
The next issue to be addressed was which computer programming language should be used for this exercise. Clearly any number of programming languages would be suitable, ranging from BASIC to C++. Some of these programs have steep learning curves, and not all could be obtained for free by the students. It was decided that they should use the programming language used by many bioinformaticists—Perl (Practical Extraction Report Language; see www.perl.com).
There were several reasons to use Perl for this assignment. It could be obtained without cost for either the Macintosh or the PC (Table 1). It would be reasonably easy for students to learn enough Perl to accomplish the assigned task, and there are numerous resources on-line for learning Perl. One of the key arguments made for Perl's suitability for this and other programming projects is that it is a good language “for getting things done.” It seems likely, however, that other comparable programming languages such as Python ( www.python.org) and Ruby ( www.ruby-lang.org/en/index.html) could also be used for this purpose. Perl, Python, and Ruby are all used in open source efforts to develop bioinformatics software ( www.bioperl.org).
The programming assignment was described for the students at the beginning of the semester, and from the information in the surveys the two students with prior programming experience were asked to be the leaders of the two groups. The team leader and the group members would work together to design, code, and test the program. Early in the semester the goal of the program was discussed, and the class worked out the basic logic of how the program would achieve this goal. It was described how the program could first be developed in the form of pseudocode—a detailed English language representation of what a program must do—and then, with the team leaders' help, the program would be coded into Perl. The importance of the full participation of the group was stressed. It was stressed that programming is ultimately an exercise in clear and logical expression of the means by which a particular goal is achieved—and that all of the students, even those lacking computer programming experience, had the ability to participate in this aspect of the program's development. The particulars of how the code would actually be written in Perl were worked out by the two team leaders, the individuals who had the most experience with the process (and frustrations) of coding.
Figure 1. A flowchart showing the overall flow of information in the amino acid composition calculator Perl program.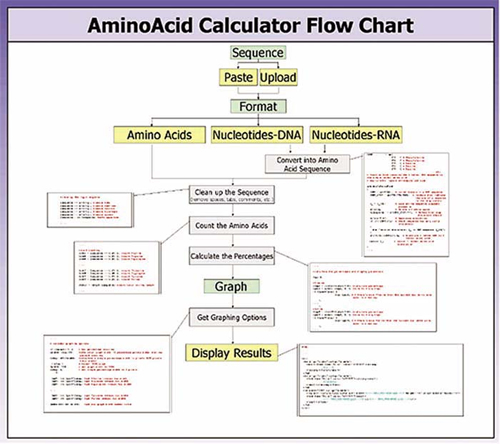
Each group began their planning, and in order to expedite their progress a working version of the program was demonstrated by the instructor. This resulted in one of the most surprising statements that this instructor has heard since he began teaching: “We can do better than that!” Within a few weeks, each group had a working version of the program, different in detail from that written by the instructor. A flowchart from one of the two groups illustrates the overall organization of their program (Figure 1), and an excerpt from their program is shown in Figure 2. It was clear from their relatively rapid progress that learning to code in the Perl language was not an impediment to progress, even though neither of the team leaders had prior programming experience with this particular language. One remarkable aspect of their programs' development was that they themselves identified and used a key feature of the language (the use of so-called regular expressions) that greatly shortened the work of tabulating the numbers of the individual amino acids. This is consistent with the Perl philosophy of programming that laziness is actually a virtue!
Figure 2. An excerpt of code from the Perl program for the amino acid composition calculator.
The program's specifications were outlined at the beginning of the semester, but later the students were offered an opportunity to obtain extra credit in this project if they implemented it as a program accessible over the Web, from a form-containing Web page. A user would copy-and-paste the sequence of a protein (or upload it from a local text file) into a text field on a specific Web page (illustrated in Figure 3). Upon clicking the submit button, the data would be sent to a Web server hosting their program, and the program (a Perl script) would receive the data as an input. Following the execution of the program, a new Web page with the results of the amino acid composition calculation was returned to the user, either represented as a table or graphically represented as a bar chart (as shown in Figure 4). By the end of the semester the students had done just that, greatly exceeding this instructor's expectations.
The final versions of each program were thoroughly tested and found to yield the expected results. Therefore at the end of the semester, the two groups had come up with two independent solutions of the problem posed in the assignment.
This fits well with the stated philosophy of the Perl programming language:“ There's more than one way to do it.” One of the team leaders, Srdan Kobsa, describes the process of the development of his group's program in the Appendix to this article.
Figure 3. A screenshot of the Web page containing the form used to submit protein sequences for analysis by the Web server-based Perl script for calculating amino acid compositions. The amino acid sequence for the ryanodine receptor (RYR1) protein has been pasted in the text field. The program depends on the sequence being in FASTA format.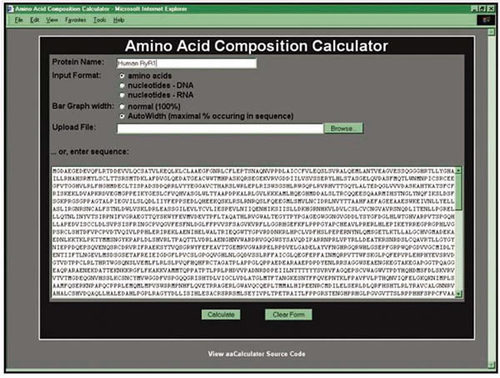
Despite this apparent success, there were problems that must be solved before this type of assignment is used again. These problems were made apparent by discussions with the students and in reading the course evaluations. These problems and possible solutions are described in the next section.
CRITICAL ASSESSMENT OF THESE EFFORTS
Several types of assessment have been employed to assay the success of these efforts including in-class and take-home examinations, a lab practical examination, and individual and group research project presentations and reports, as well as end-of-semester student evaluations of the course. Taken collectively over the last 7 years, these assessments indicate that some success has been achieved in realizing the goal of giving students practical experience in the use of these bioinformatics tools and databases. However, much more work needs to be done to see that students think about the biological meaning of the results of their analyses.
In lieu of an in-class final examination, the students in the bioinformatics course were given a take-home examination to assess their individual abilities at the end of the semester. This examination consisted of a focused set of questions to be addressed using tools and concepts previously covered (see Table 6). Nearly all of the students that passed the course showed evidence from their individual examinations that they had mastered specific technical skills used in bioinformatics (Table 7). The critical question is whether these hard-won skills had resulted in the development of increased insight into the biology of the systems at which the students were looking. This is a harder and more subjective assessment. With regard to this point, the students clearly differed in their ability to understand and articulate the biological significance of the results of their bioinformatics studies. This is, of course, a far more valuable assessment than whether the students say that they enjoyed the course or whether they say that they had learned a lot about bioinformatics during the semester.
| Using a combination of computer and Internet-based tools, you should write a minireview that summarizes what is known about the structure and function of each of the protein or RNA molecules associated with (1) the vault ribonucleoprotein and (2) the apoptosis-inducing factor (AIF). You should also briefly describe the biological and medical relevance of these findings. |
| Address the specific issues associated with each of the two topics. |
|
| Skill | Proportion of enrolled students demonstrating technical ability to perform specified task |
|---|---|
| Find required sequences in GenBank | 9/11 |
| Generate multiple sequence alignments using ClustalW | 10/11 |
| Identify conserved domains in proteins | 9/11 |
| Predict secondary structure for RNA molecules | 10/11 |
| Model homology via Cn3D | 10/11 |
Figure 4. A screenshot of the Web page containing the results of the amino acid composition analysis carried out by the Web server-based Perl script. In this version, the analysis is returned in the form of a bar graph generated through the use of a specific Perl module (HTML::Template).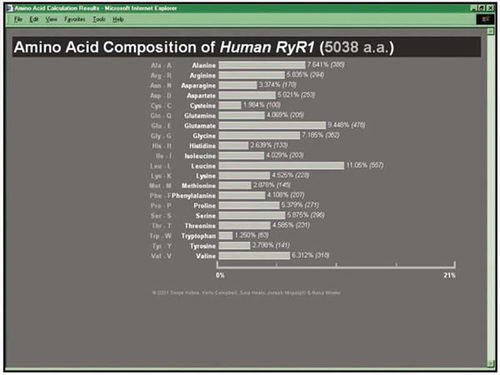
Other observations of students at work in the computer laboratory have highlighted the need for an instructor to directly confront this problem. When working on the exercises in the computer laboratory, some students were observed to rush through the individual steps of an exercise, spending little time thinking about the meaning of the results they obtained along the way. Although they quite successfully followed the instructions for these exercises to completion, they were occasionally unable to verbalize what insight they have gained from their analysis or how the result obtained related to what they have learned in previous course work. For other students, with weaker computer skills, the technical challenge of completing the exercises also served to distract them from thinking about the meaning of their results.
What can an instructor do to address this problem? First, it is important to recognize that some undergraduate students confuse technical mastery with insight. These students think of their ability to list facts, to carry out a procedure, to make calculations, or to run a computer program as being equivalent to understanding in a discipline. On examinations, they may write about the words that make up a question without ever addressing the specific question being posed. It is not always easy to convince these students that there is a difference between a listing of facts or observations and drawing inferences from those facts or observations.
One way to address this persistent problem is for the instructor to take students step by step through several specific examples of how the results of bioinformatics studies can lead to biological insights. It was clear from in-class observations that most students can quickly learn to perform the specific computational tasks required for bioinformatics studies. In the classroom the instructor should focus on the interpretation of the results, instead of the methods by which the results were obtained. Fortunately for instructors, textbooks are now available that take just this sort of approach to the study of bioinformatics (e.g., Campbell and Heyer, 2002).
The course evaluations from the bioinformatics course, as well as conversations with individual students, highlighted another problem that needs attention. The majority of the students indicated on their course evaluations that they were unhappy with respect to their participation in the programming project. Even though these evaluations were anonymous, it was clear that this dissatisfaction emanated from the members of the group other than the team leaders. The students were instructed in the guidelines for this assignment that “everybody is to be involved in the design, development, and testing of the [Perl] program.” Despite the overall success of the groups in completing the projects, it was clear that this did not take place in the way that this instructor had intended.
The obvious solution in the future is that during the course of a programming project, individual students will be given specific assignments to complete, for which they will be accountable, to both their fellow group members and the instructor. It should be emphasized that there are many aspects to a programming project that do not involve the direct coding of an algorithm. Students could be assigned to specific tasks such as documentation of the program's features, annotation of the source code, testing of the program with a variety of data (including nonsensical data), and validation that the program reports the correct result for any input data. Another important task for a student in the group could be the evaluation of the user interface of the Web pages used for data submission or the Web page used to report the data analysis. All of these activities represent the work of individuals or divisions in companies that develop commercial software packages.
Although there is clearly room for improvement of the implementation of these courses, there is also evidence that these courses have provided some of the students with a working knowledge of bioinformatics that they subsequently put in practice in the research laboratory. A student recently sent the following note: “I wanted to let you know that I have used a lot of what I learned in your classes to do my work [a summer research internship], which has been entirely molecular methods! My mentor was impressed that I had experience using many bioinformatics programs that he uses himself.” This is the kind of feedback that makes these efforts worthwhile.
SOME FINAL OBSERVATIONS AND SUGGESTIONS
A number of observations and suggestions come to mind about the practical aspects of developing and running courses that include bioinformatics content. These issues and some possible solutions are described below.
Computer Laboratory Problems. Ideally an instructor will have access to a dedicated computer laboratory for the first part of the semester. It is important that the instructor be directly involved in the installation, configuration, and testing of the required software packages on these computers, in order to ensure that the exercises can be carried out by the students without unnecessary difficulty. Unfortunately this is more difficult if an instructor is using university-wide computer labs where he or she will have to effect the installation through the college or university's IT staff. In the latter case, it is important to give the IT staff adequate lead time for installation so that the instructor can make sure that the laboratory time with the students will not be unnecessarily wasted on time devoted to troubleshooting software or hardware problems.
Student Complaints. A common complaint heard from some students in the advanced courses is that this course work takes too much time. More often than not, this is because some students completely underestimate the amount of time it takes to carry out computing tasks. It may seem counterintuitive to some students, but it is actually possible to become more efficient through lots of practice at the beginning of the course. The more students do this work, the more likely they will find efficient ways to carry out the procedures. With practice it is also possible for them to avoid many of the pitfalls that can consume large amounts of time.
Need to Establish Checkpoints in Research Projects. Students often get behind on individual or group projects for the reason stated above. As a result the quality of their final presentations or papers suffers. Consequently the particular portions of the research are now assigned to be due on specific dates. Meetings are set up with the students at regular intervals to look for concrete evidence of progress. In addition, it is important to leave sufficient time for students to synthesize their research findings into a final paper and presentation at the end. When these changes have been implemented they have resulted in clearly improved quality of the students' final submitted work. Some of these reports were of sufficient quality that they could be edited and submitted for publication in one of an increasing number of undergraduate research journals.
Do Not Give Them Too Much Help. It is both reasonable and necessary to come to the aid of students when they are bogged down in a problem associated with carrying out these exercises. However, it is also imperative that students learn how to solve some of these problems for themselves. An instructor occasionally finds himself or herself in a situation where students are requesting lots of help for every single computing task they find difficult. It is important to encourage the students to first read the supporting documentation for software programs in order to find the answers for themselves. It is vital to resist the temptation to“ rescue” the floundering student too soon.
Get a Good Sense of Group Dynamics. It is important to check that work on group projects is equitably distributed among the members of the group. Nothing makes for ill will in a student group more quickly than the perception that some of the students are not doing their fair share of the work. It is also important for the instructor to look out for potential personality conflicts. Some students who decide to work together at the beginning of the semester soon find themselves at odds with each other. Personality conflicts all too often reduce the effectiveness of the learning opportunities available to students through collaborative research projects.
Enlist the Help of Computer Science and Mathematics Faculty. If an instructor wants to include a programming project in a course, it makes sense to enlist the help of the local computer science faculty—they have the pedagogical experience in this area. Similarly, a mathematics faculty member could be very helpful in getting students to understand and appreciate the aspects of probability and statistics relevant to bioinformatics. Many of these faculty members are very interested in finding ways to apply their extensive experience to the problems posed by other scientists, including biologists. There is abundant evidence that these sorts of collaborations can be very productive for everyone involved.
SOME FUTURE GOALS
In addition to implementing the improvements mentioned above, there remain many exciting opportunities to develop new exercises that reflect the growth and development of bioinformatics as a discipline and the ways in which it can contribute to biology and medicine. It has been suggested that the students could use on-line tools to analyze the gene makeup of a relatively small genome, such as those of bacteria or viruses.
It seems reasonable now that a programming project could be a regular aspect of the bioinformatics course. The publication of several recent books that introduce the programming aspects of bioinformatics will facilitate these efforts. In the future it would also be useful to have students not only develop new programs, but also improve and extend the capabilities of programs created by previous classes.
One of the things that this author wants to do in the future in these courses is to spend more time talking about what has been discovered. Just a discussion of the discoveries noted in the papers describing the first draft of the human genome could easily fill a semester. Clearly we are still at the very beginning of an exciting era of biology, and bioinformatics will contribute heavily to the expansion of our understanding of the molecular basis of life in the years ahead. We must envy the students that are just starting their careers in biology!
APPENDIX
A STUDENT'S PERSPECTIVE ON PROGRAMMING A SIMPLE BIOINFORMATICS APPLICATION IN PERL
By Srdan Kobsa
Much like any computer application, developing an amino acid composition calculator presented several challenges. The primary goal of any well-designed computer application is its ability to solve a particular type of problem. Furthermore, the best measure of the program's success in achieving this goal is the user's evaluation. To be successful, a 3D-design application must be useful to an architect. A program that calculates the percentage composition of amino acids in a protein sequence should be designed for a molecular biologist. Regardless of how elegant the computer algorithm is to a computer scientist, if the final product makes no sense to a biologist, it has failed. A good program does not assume or require the end user to understand the underlying computer logic and language.
This, however, brings up a crucial requirement: It makes it necessary for the person creating the program to understand how the final user will use the information. This is not always trivial, considering that, for example, a biologist's understanding of a particular problem might be very different from the approach that the programmer will have to adopt in order to make the computer carry out a particular task. The development of our program required the point of view of individuals with background in both biology and computer science. Since members of our group possessed two perspectives, we were in a good position to tackle this problem.
The programming project was divided into distinct phases. Each phase represents a step from the biological problem toward the computer logistics of the solution. Naturally, the first step was planning. The students collaborated to develop a clear idea of the goals of the project and the specific approaches and functions the program was to have. A simple and widely used sequence format, FASTA, was chosen as the default input ( www.ncbi.nlm.nih.gov/BLAST/fasta.html). The user was to enter the name of the protein followed by its amino acid or the in-frame nucleotide (DNA or RNA) sequence in FASTA format. If needed, the program would translate the nucleotide code into the amino acid code and then calculate the percentage of each amino acid in the deduced sequence. The user would then be presented with the results. This report would include the name of the protein being analyzed, its total length, and both the number and the relative percentage of each of the 20 amino acids. This represented the most challenging aspect of the program that was to be coded. The results would be represented graphically by horizontal bar graphs corresponding to the percentage of each amino acid used. However, since the percentages of the 20 amino acids can be quite low (a single amino acid rarely composes more than 50% of any single protein), an option to view the graph on a smaller scale was added. In other words, instead of using the 0%–100% scale, a user would be given an option to be presented with a graph on a scale from the zero percentage up to the maximum occurring percentage of any amino acid in that specific protein (e.g., if a protein contains the most alanine—37.8% then the graph would be scaled from 0 to 45% instead of 0 to 100%, making the bars more distinct in length). This option was added purely for visual benefittothe user.
By far the most practical and accessible way to implement these functions is the World Wide Web. In fact, a myriad of computational biology tools already exists on the Web, ranging from simple ones such as this amino acid composition calculator to complicated structure prediction algorithms. The program would be accessible to anyone in a very simple and universal format. In addition to the HTML used to compose the Web elements needed for the user interface, the actual computational operations were carried out in Perl, which is, traditionally, one of the most commonly used programming languages on the Internet.
The project was now ready for the second phase—the development of the details of the programming algorithm in the form of a flowchart (Figure 1). This represented the transition between the practical reasoning of biology and the computer logic. The information flow in the amino acid composition calculator starts with the user entering the name and the sequence of the protein. The user also specifies whether the entered data are an amino acid, a DNA, or an RNA sequence and selects a particular type of graph. Those inputs are then sent by the computer over the Web to the server hosting the Perl program to be processed. First, the sequence is checked. If an empty input or nonallowed characters are encountered, an error is reported. Next, the identifier line included in the FASTA format is removed, if one is present. Any additional characters, such as spaces, tabs, line feeds, and returns, are also checked for and removed if present. Finally, the remaining letter sequence is made uppercase (for uniformity). If the entered sequence is DNA or RNA, it is translated into the 20 single-letter amino acid code using the standard genetic code. The number of letters in this final character string is then counted. It represents the total length of the protein in amino acid residues. Next, each of the 20 individual amino acids is counted by summing up the number of times the particular amino acid code letter appears in the sequence. The ratio of the two values gives the relative percentage of the specific amino acid in the sequence. If the user has selected the option to scale the graph according to the maximum percentage that appears in a particular query, the largest percentage is also recorded. Finally, a separate Web page containing the results is generated.
The final stage of the project consisted of actually writing the computer code (see excerpt in Figure 2). Students that had more extensive experience in computer programming and Web page design were especially involved in this stage. A Web page containing a submittable form was constructed, as this would be the way by which the user would enter the information and select desired options (Figure 3). This Web page was linked to a Perl executable script in a way that took the information entered by the user and assigned it to computer variables. The text variable containing the actual sequence was key. A Perl function (length) was used to evaluate the length of the string. If it was found to be zero, indicating an empty sequence, an error was reported. A function that replaces characters in a string (s///) was used to remove spaces, tabs, line feeds, and returns. Each of these was simply replaced with an empty character (not a space). Finally, the uc function was used to make the whole string uppercase. If needed, an array was defined containing the amino acid dictionary, then used to translate nucleic acid codons into amino acids. To count the particular letters in the string, a function that looks for a specified string and then returns the number of occurrences was used (tr). A temporary variable was also assigned to contain the maximum number of occurrences of any amino acid. Each time an amino acid was counted, the number was compared to the current maximum in the temporary variable. If the new count was larger than the current value of the temporary variable, the maximum was replaced with the new number. This made it possible to evaluate the maximum scale value on the bar graph.
In order to use HTML with Perl, a special module (HTML::Template) was downloaded form the ActiveState Perl Web site ( www.activestate.com). This module made it possible to include Perl-generated variables into ordinary HTML. The bar graphs were developed using a very common “trick.” Having a solid color image of a fixed height makes it possible to simply stretch it in HTML (<img width=”X”> variable) to a desired width. The widths were calculated by simply multiplying the percentage of a particular amino acid with the set width of the bar graph. For example if the percentage of leucine was found to be 8.7% and the width of the full bar graph (100%) was set at 500 pixels, the width of the leucine bar would be 500 × 0.087, or 43.5 pixels. Since the image width in HTML has to be an integer, a Perl function (int) was used to round the numbers. In the case that the user had selected the maximum occurring percentage as the scale for the graph, that percentage (increased 10 percentage points) would be given a value of 500 pixels, and all the other math would be adjusted accordingly (e.g., if the full length of 500 pixels represented 45%, then 8.7% for leucine would equal 97 pixels). Once generated, the resulting Web page was sent to the user's Web browser as a response to the query (Figure 4).
This project not only involved extensive teamwork, but required drawing upon distinctively different areas of expertise of students in the group. It revealed both the issues and the connections between computational biologists and software engineers. While small in functional scope, this project served as a very good model, which remains open to additional development.
FOOTNOTES
Monitoring Editor: Erin Dolan
ACKNOWLEDGMENTS
I would like to thank the many students of the cell biology, structural biology, and computational biology courses for their help in developing and refining the exercises described herein. I express gratitude to Srdan (Serg) Kobsa for his contribution to this paper. I thank the reviewers for their constructive critiques of the manuscript. I would also like to thank A. Malcolm Campbell and Donald Stratton for their help in thinking about student assessment.



