Students’ Understanding of the Dynamic Nature of Genetics: Characterizing Undergraduates’ Explanations for Interaction between Genetics and Environment
Abstract
The idea of the interaction between genes and environment in the formation of traits is an important component of genetic literacy, because it explains the plastic nature of phenotypes. However, most studies in genetics education characterize challenges in understanding and reasoning about genetic phenomena that do not involve modulation by the environment. Therefore, we do not know enough to inform the development of effective instructional materials that address the influences of environmental factors on genes and traits, that is, phenotypic plasticity. The current study explores college students’ understanding of phenotypic plasticity. We interviewed biological sciences undergraduates who are at different stages of their undergraduate studies and asked them to explain several phenomena that involved phenotypic plasticity. Analysis of the interviews revealed two types of mechanistic accounts: one type described the interaction as involving the environment directly acting on a passive organism; while the other described the interaction as mediated by a sensing-and-responding mechanism. While both accounts are plausible, the second account is critical for reasoning about phenotypic plasticity. We also found that contextual features of the phenomena may affect the type of account generated. Based on these findings, we recommend focusing instruction on the ways in which organisms sense and respond.
INTRODUCTION
Over the past several decades, scientific advances in genetics have enhanced our understanding of the complex underlying mechanisms of genetic phenomena. Ripples from knowledge generated in research labs have reached the public sphere and are impacting citizen decision making about many issues, including personalized medicine, gene therapy, genetically modified crops, forensics, and genetic screening. It is, therefore, well accepted that learning genetics is a crucial component of scientific literacy (Boerwinkel et al., 2017).
However, research has shown that this domain is challenging for both teaching and learning for several reasons. First, many of the core entities and processes in genetics, particularly molecular genetics, are unfamiliar to high school and university students (Marbach-Ad and Stavy, 2000; Lewis and Kattmann, 2004; Duncan and Reiser, 2007). Second, genetic phenomena transverse multiple levels of organization, and reasoning across these levels is challenging for students (Stewart et al., 2005; Gericke and Wahlberg, 2013; van Mil et al., 2013; Reinage and Speth, 2016; Haskel-Ittah and Yarden, 2017). A third challenging aspect of genetic phenomena that has not received much attention in genetics education research is the complex interactions between the environment and genetic mechanisms in the development of traits.
In the post-genomic era, there has been a shift within the scientific community from mere acceptance of the mutual effect of nature and nurture to a more complex understanding of gene–environment interaction (Darling et al., 2016). While the genome serves as the background programming, so to speak, the environment modulates the outcome, that is, it interacts with this programming to influence the phenotype. This modulation of the trait is interactional, involving complex interactions between genes and the environment, rather than simply additive (Moore and Shenk, 2017).
Understanding that phenotypes are the developmental result of interactions between genes and environment is not only important for scientists and scientific inquiry. It is also important for personal and civic engagement with genetics issues. A recent consensus study of experts’ views of what constitutes critical ideas for genetics literacy identified the idea that “multiple genes and multiple environmental factors interact in the development of most traits” as potentially the most important aspect of genetic literacy (Boerwinkel et al., 2017, p. 1105). Yet these interactions between genes and the environment are largely overlooked in biology textbooks, curricula, and instruction. Omissions of these interactions and the oversimplification of inheritance by focusing on monogenic traits (Dougherty, 2009; Dougherty et al., 2011) lead to problematic deterministic views of genetic traits as being predominantly decided by genes rather than being modulated by both genes and the environment (Castéra et al., 2008; Gericke et al., 2014; Puig and Jiménez Aleixandre, 2017).
To some extent, the simplification of genetic phenomena and their mechanisms is necessary to make these ideas accessible and understandable to students. However, such simplifications run the risk of engendering genetic determinism and essentialism. These are serious obstacles to a productive understanding of genetics, because they obscure the probabilistic and developmental nature of genetic traits (Jiménez-Aleixandre, 2014; Todd and Kenyon, 2016; Stern and Kampourakis, 2017). Studies show that, when students learn about Mendelian traits, which are presented in a deterministic manner, or learn about genes for certain diseases, they may develop more racist views (Donovan, 2014, 2016; Jamieson and Radick, 2017).
Such conceptions of genetics make fertile ground for stereotyping and prejudice (Keller, 2005). This is because conceiving of genes as the direct (and unmediated) cause of traits may lead to views of socially constructed human differences (gender, race, sexual orientation) as immutable, stable, and discrete when they are not (Dar-Nimrod and Heine, 2011). Studies of biology curricula and textbooks show that the ways in which they represent traits and their causes often contribute to deterministic and essentialist views rather than reducing them (Donovan, 2014, 2017; Jamieson and Radick, 2017). This, in turn, fosters prejudice and contributes to sustaining social discrimination (Donovan, 2017). Hence, genetic determinism is an educational problem with serious social implications.
Understanding the mechanisms of interaction between the environment and the “genetic program” can help mitigate against genetic determinism, because it makes salient the role of the environment in influencing phenotype and trait variation. However, understanding these mechanisms is not trivial. First, because laypersons tend to assume an additive contribution of the environment to the trait (Moore and Shenk, 2017). This conception is the result of misapplying the contribution of the environment to variation in traits at the population level to the individual level (Moore and Shenk, 2017). For example, a population-level study may suggest that the variation in height in a certain population could be explained by 90% genotypic variation and 10% environmental variation. This can be misinterpreted by applying it to the individual level, and supposing that 90% of an individual’s actual height is the result of genetics and that 10% can be attributed to nutrition or other lifestyle factors. Such a view suggests a simple additive model of “gene + environment = trait” that contradicts the scientific view of how genes and environment interact at the individual level (Moore and Shenk, 2017).
Second, there are multiple mechanisms by which the environment may affect an individual’s traits. These mechanisms act at different levels: physiological reaction, gene regulation (including epigenetics), and mutations (Yona et al., 2015). There is also a crucial difference between the mechanisms acting at these three levels. In the case of mutation, the environment affects the phenotype by changing the DNA sequence. Whereas at the other two levels (physiological and gene regulation), there is a response of the organism to the environmental change by activating mechanisms that modulate the trait. This modulation is termed phenotypic plasticity.
Batzli and colleagues (2014) found that more than 60% of university-level students were not familiar with the concept of phenotypic plasticity as opposed to 5% who were not familiar with the concept of phenotypic variation. In addition, these students highly associated the word “mutation” with the concept of genetics, while the term “phenotypic plasticity” was not associated with genetics at all (Batzli et al. 2014). Acknowledging only mutagenic effects of the environment is problematic, because it still engenders a deterministic view of genetics (once the gene is altered it still solely determines the phenotype). Hence, if we are aiming to reduce deterministic conceptions, we should focus on phenotypic plasticity as a core set of mechanisms in biology.
Most genotypes exhibit phenotypic plasticity to some extent, whether a phenotype is defined as a developmental event (e.g., changes in the development of gills in certain fish as a result of water oxygen levels), a physiological adjustment (e.g., tanning), a behavioral shift (e.g., changes in locusts’ behavior to solitary or sociable dependent on the density of individuals), or an environment-dependent gene expression (e.g., the lac operon response in bacteria; Kelly et al., 2012). These mechanisms involve signals from the external or internal environment that lead to biological, genetically based responses (DeWitt and Scheiner, 2004) at different regulatory levels (e.g., inhibiting/activating proteins, affecting RNA translation or transcription). Their resulting effects on the phenotype may be transient or may persist through cell divisions (in the case of epigenetics) and may even be passed on to the organism’s offspring (Yona et al., 2015). Hence, although the environmental conditions triggered the change, it is the organism’s responsive and regulatory mechanisms that bring about the phenotypic response.
Returning to the example of an individual’s height, the amount of nutrients is sensed, and consequently, growth hormone production or secretion is modulated, thus altering growth and other processes. In the case of malnutrition, at first, biological mechanisms are altered via transient short-term mechanisms (e.g., protein modifications or regulation of gene expression). When malnutrition persists, an epigenetic mechanism can be triggered to permanently turn growth-related genes on or off.
Epigenetic modifications occur in different ways, and one prominent mechanism is the addition of methyl groups to specific DNA bases, resulting in blocking of gene expression. In some cases, the methylated DNA can be transferred to the next generation and may affect the phenotype of the offspring, even if their own nutrition is adequate (Martins et al., 2011).
As mentioned earlier, understanding the mechanisms by which phenotypic plasticity occurs is important not only because it reflects current knowledge in the field of genetics, but because it may also serve other educational goals. Arguably, understanding that genes are not the ultimate (or only) determinants of traits, but that environmental signals can lead to the modulation of traits (not via mutation), may help reduce students’ tendency toward genetic determinism. In addition, understanding the plasticity of development establishes the ground for dealing with microevolution processes and evo-devo (Kampourakis and Panagiotis, 2018).
While the importance of avoiding a deterministic conception of genes and emphasizing their plastic nature is acknowledged by science educators, it is not clear how this complex idea should be taught. Boerwinkel and colleagues (2017) suggested that understanding that gene expression is regulated (i.e. knowing that different genes are switched on and off in different cells and at different times) is sufficient for a 21st-century genetic literacy, but that knowledge about the specific mechanisms by which environmental signals lead to this switching is not necessary. This sentiment is echoed in research on genetics learning progressions (Duncan et al., 2009; Elmesky, 2013). For example, Duncan et al. (2009) state that: “We are not advocating here that students need to understand the mechanisms of gene expression and how this biological process is regulated, rather we argue that students need to understand that most cells in a given organism have similar genetic content but that the usage of this content is regulated in time and place” (p. 662).
In the absence of research, it is not clear whether providing information that 1) both genes and the environment modulate our traits and 2) genes can be turned on and off would be enough for students to be able to explain how genes and environment interact at the individual level and to reason about phenomena and issues they are likely to encounter as citizens. In fact, recent studies suggest that this information, by itself, will not be enough (Gericke et al., 2017; Puig et al., 2017). These studies have shown that knowledge about the mechanisms of gene expression and gene regulation does not directly lead to the ability to explain phenomena involving regulation of gene expression as a result of environmental signals (Puig et al., 2017).
In summary, while understanding the mechanisms of phenotypic plasticity is important, it is also challenging for students. We currently do not know much about how students reason about phenotypic plasticity, or what ideas they bring up when trying to explain these phenomena. Therefore, our current study aims to characterize the kinds of mechanisms students propose when explaining phenotypic plasticity phenomena. We take into consideration that students’ accounts may be influenced by their prior knowledge and contextual features of the phenomenon (Duncan, 2007; Nehm and Ha, 2011). Therefore, our research questions are:
What kinds of mechanistic accounts do undergraduates provide to explain how environmental conditions influence genetic traits?
How do contextual features of these phenomena influence the types of accounts generated?
THEORETICAL FRAMEWORK
Mechanistic Reasoning and Agency of Change
Since Galileo and Newton, natural phenomena have been explained by underlying mechanisms (Westfall, 1971). Thus, supporting students’ ability to construct mechanistic explanations is one of the core objectives in science education. This, however, is a challenging goal, because mechanistic reasoning entails connecting between multiple levels and multiple causes (Wilensky and Resnick, 1999; Russ et al., 2008; Krist et al., 2018) and requires some level of domain-specific knowledge (Duncan, 2007).
In the past several years, studies in philosophy of science and science education have tried to sharpen the definition of what counts as a mechanistic explanation (Darden, 2008; Craver and Darden, 2013; Russ et al., 2008; Krist et al., 2018). These studies suggest that a mechanistic explanation should include entities, their properties, activities, and interactions (Russ et al., 2008; Craver and Darden, 2013). The interactions between the entities should be linked in a way that explains how they lead to the explained phenomenon (Krist et al., 2018). While these studies characterized the components of mechanistic explanations, they did not address what kinds of understandings are needed to decide which entities, activities, and interactions are relevant and which are not.
Based on our own prior work, we propose that these decisions depend on domain-specific knowledge of appropriate entities (and their activities) in the domain (Duncan, 2007; Haskel-Ittah et al., 2019). For example, when we asked seventh-grade students to explain the effect of genes on certain traits, they used the entity “protein” only in cases in which they knew of a relevant protein activity that could account for the formation of the trait in question. If they could not think of a relevant protein activity, they did not invoke that entity at all in their explanations. Thus, ideas about proteins and their activities (or functions) are essential to developing plausible mechanistic accounts of genetic phenomena. Hence, it seems that generating mechanistic accounts involves identifying relevant entities and their relevant activities to explain the target phenomenon (Haskel-Ittah et al., 2019).
In addition to selecting relevant domain entities and activities, there is also a process of deciding which of these are most central to the mechanism. This decision process occurs both in scientific explanations and everyday explanations and is dependent on the goal of the explanation (Craver and Darden, 2013, pp. 33–34). Not all entities and activities that are somehow involved in the mechanism would be (or should be) mentioned in an explanation, because not all are relevant to the purpose of the explanation. The more complex the phenomenon, the more “pruning” of entities and activities needs to be done by the individual giving the explanation in order to keep it fruitful and parsimonious (Cheng and Novick, 1991). One prominent part in the process of deciding which entities are important enough to appear in the explanation is the identification of the agency of change (Murayama, 1994). Entities and activities whose existence directly and uniquely caused a particular outcome reflect the agency of change for that outcome. For example, a mechanistic explanation aimed at explaining a plane crash would focus on the entities involved in a technical failure rather than on gravitational forces. Gravitation is, of course, one of the causes for the crash, but when comparing the crash to normal flights, gravitational force existed in both outcomes and thus did not lead to the difference between the two cases (Cheng and Novick, 1991). Hence, in this case, a wire causing a short circuit is uniquely and directly involved in the outcome and is thus one of the agencies of change, whereas gravitons (hypothetical quantum of gravity) are not (Murayama, 1994).
Mechanisms of phenotypic plasticity are complex and involve multiple entities and interactions. Therefore, it is reasonable to assume that, when asked to explain a change in phenotype, students would not provide an explanation that includes all possible entities and interactions. Instead, they would most probably focus on the entities that they perceive as more central and that are likely more familiar to them. Thus, an analysis using the perspective of agency of change and domain-specific knowledge of entities and activities, would allow us to identify which entities, activities, and interactions are invoked as both plausible and central to phenotypic plasticity phenomena in students’ explanations.
The Effect of Contextual Features on Reasoning
In considering what ideas students bring to bear as they construct a mechanistic explanation, we take into account the contextual features of the phenomenon that may lead to the elicitation of those ideas. Current conceptions of knowledge, intuitive or learned in school, suggest that it exists as fragmented pieces that may be activated by specific contextual cues and compiled into an explanation in specific situations (DiSessa, 1993; Kapon and diSessa, 2012). This context specificity of knowledge may lead to the appearance of different kinds of ideas and explanations of essentially the same phenomenon when presented in different contexts (Clark, 2006).
This contextual effect in reasoning was also shown in biological domains such as ecology, genetics, and evolution (Kampourakis and Zogza, 2009; Nehm and Ha, 2011; Shea et al., 2015). For example, Nehm and Ha (2011) found that students used more naïve ideas and fewer scientific ideas when explaining evolutionary processes in the context of trait loss than when explaining it in the context of trait gain. Similarly, Shea et al., (2015) found that the contextual feature of organism type impacted the level of sophistication of students’ responses. Students provided more complex explanations in the context of problems involving human genetics compared with plant genetics.
In this study, we aimed to characterize students’ accounts of phenotypic plasticity phenomena in contexts that differed in terms of the organism involved and the timescale of the phenomenon (transient to cross-generational). As part of our analysis, we therefore explored how these contextual features of phenotypic plasticity phenomena impacted students’ accounts (research question 2).
Our study involved students at the university level, because prior research suggested that younger (high school) students may not have sufficient domain-specific knowledge to generate appropriate accounts of such complex genetic phenomena (Marbach-Ad and Stavy, 2000; Haskel-Ittah et al., 2019). We therefore interviewed university undergraduate students whom we thought might be more familiar with interactions between genetics and the environment (Duncan, 2007).
METHODS
Data Collection
We conducted a cross-sectional study with undergraduate students in their first, second, and final year of a major in a life sciences–related field. Students were recruited through an invitation in a students’ Facebook group. This group was established by campus students majoring in life sciences–related fields for the purpose of publishing issues related to students’ lives, including job offers and offers for recruiting students as humans subjects for various studies on campus. This group has 2700 students registered participants, which makes it ideal for reaching a broad range of undergraduates. Our recruitment post noted the (only) eligibility criterion: majoring in a life sciences–related field.
We interviewed 38 (24 female and 14 male) undergraduate students: nine first-year students, 15 second-year students, and 14 third-year (final year) students, for about 45 minutes each; all interviews were recorded. Pseudonyms are used in place of students’ names in examples provided in the result section. All the students were studying toward a degree in a life sciences–related field (nutrition, animal sciences, biochemistry). While these majors do differ, they share some similar basic course requirements. Therefore, all of our students completed a basic cell biology course, and all of our second- and third-year students completed a basic genetics course (described later). Some third-year students we interviewed also completed an advanced genetics course. The syllabus of the cell biology course included the topics of: cell structure, the central dogma and protein synthesis, enzymes, DNA structure and the genetic code, and the cell cycle. The syllabus of the basic genetics course included Mendelian genetics, the cell cycle, cytoplasmic inheritance, mutations, and genetic mapping. The syllabus of the molecular genetics course included genetic engineering, alternative splicing, reverse genetics, transcriptional regulation, and epigenetics.
During the interviews, students were asked to explain four genetic phenomena (the full descriptions of the four tasks are available in the Supplemental Material):
Tanning: UV exposure leads to a darker skin.
Stunting: Malnutrition in children leads to reduced growth rate, thus causing them to be shorter than what would be expected based on their age and parents’ height.
Trans-generational stunting: This case was based on the previous one but included additional information about the offspring of stunted children who also exhibit reduced growth rate despite receiving adequate nutrition.
Smell imprinting: Caenorhabditis elegans worms exposed to food with a particular odor are attracted to this odor and lay eggs as a result of sensing this odor. Once exposed to the odor, worms persist in this response, even when there is no food present. Interestingly, their offspring are also attracted to the odor and increase egg-laying activity, even if they have never been exposed to the food in conjunction with the odor.
Each of the four tasks represents a case of phenotypic plasticity. All cases involve a genetic trait that is modified in response to an environmental change. While these tasks all involve the similar underlying idea of phenotypic plasticity, there are some contextual features that differ among them: 1) the organism (tasks 1, 2, and 3 are in the context of human traits, and task 4 describes a phenomenon in nematodes); 2) the stimulus: in each case, the environmental stimulus for the phenotypic change is different (UV in task 1, lack of nutrients in tasks 2 and 3, and odor in task 4); 3) the duration of the effect: in each task, the persistence of the phenotypic change is different (short, days in task 1; intermediate, lifetime in task 2; and long term, multiple generations in tasks 3 and 4). Interviewees were directed to provide a plausible mechanistic explanation of how the environmental stimulus led to the observed phenotype.
Analysis
Because we know little about how students explain phenotypic plasticity, the aim of this study was to map the space of the kinds of explanations students provide and their fruitfulness in providing a plausible account. By this we mean that we did not compare their reasoning with canonically accurate accounts (what an expert would say), but rather a biologically plausible one (see “Task Analysis” in the Supplemental Material). For example, without knowing how tanning occurs as a response to UV light, a mechanism in which enzymes modify melanin to make it darker, a mechanism of redistributing melanin molecules, and a mechanism of increasing melanin production are equally biologically plausible and therefore provide a satisfying account. But there is scientific evidence only for the last two (Tadokoro et al., 2005).
We define a mechanism as an explanation that includes entities with specific functions that interact with one another and lead to the emergence of the phenomenon (Machamer et al., 2000; Craver and Darden, 2013). In our analysis, we found that only a minority of the explanations were not mechanistic (4.8%, all by second-year students); namely, they did not provide any entity or activity that linked the environment and the phenotype. In these responses, students either reiterated the phenomenon without actually explaining it, explained why this plasticity is beneficial to the organism without explaining how it occurs, or indicated they did not know how to explain the phenomenon. When students did not provide a clear mechanism, they were asked again how the phenomenon occurs and were directed to provide a mechanism by being asked “How do you think it happens?” or “Explain how X [environmental factor] actually causes this [outcome] in the body?” We did not further analyze responses that did not include a mechanism at all. The remaining explanations included a mechanistic account of sorts. Some of the explanations students provided were more complex, included many entities, and explicitly mentioned how entities are linked; other explanations were vaguer and suffered from many gaps or simply reiterated the phenomenon. If students did not mention the involvement of proteins or genes, they were explicitly asked “Do you think proteins are involved in this?” or “Do you think genes are involved in this?”
As noted earlier, we first coded whether students’ answers to each task included a mechanism or not (coded as 1 or 0 accordingly). Mechanistic explanations were then further analyzed by searching for the agencies of change they posited. This was done through a constant comparison between students across tasks and between tasks for the same student. Two key themes emerged from this analysis, which we will discuss in more detail in the Results section. We labeled the two themes “direct interaction” and “sensing–responding interaction” and categorized the data accordingly. We were able to categorize all of the mechanistic explanations students provided under these themes. Students proposed either one or, at most, two possible mechanisms per question. Altogether, students could provide between zero and eight explanations (four cases, up to two explanations for each). We calculated, for each student, the number of explanations of the direct interaction type (which we termed “direct interaction score”) and the number of explanations of the sensing–responding type (which we termed “sensing–responding score”).
To further characterize these two types of accounts, we searched for the biological ideas associated with each type. We first searched student explanations for ideas known to be important for understanding genetic phenomena; namely, ideas about proteins and their functions and about gene regulation (Duncan, 2007; Duncan et al., 2011; van Mil et al., 2013; Haskel-Ittah and Yarden, 2017). In addition to these known ideas, we also searched for other ideas that were prevalent in students’ answers and searched for correlations of these ideas with a specific type of mechanistic account.
Answers were coded either as “mentioned idea X” or “did not mention idea X” if the answer did not include these specific ideas (e.g., proteins).
To explore how contextual features of the phenomena might influence the type of accounts generated, we compared explanations given for different tasks. We focused on students’ justifications for their answers to better understand which contextual features of the tasks affected students’ answers. That is, when students suggested different types of accounts (direct interaction/sensing–responding interaction) to different tasks, we focused on their reasons for why the two cases are different and whether these reasons had to do with the different contextual features. For example, students tended to assume sensing in the case of smell imprinting, because smell is a familiar sensory experience.
Ten percent of the data were coded by two independent coders (M.H.I. and G.R.D.). The overall intercoder reliability (including explanation categorization and identification of biological ideas) was 86%.
RESULTS
Two Types of Mechanistic Accounts for Phenotypic Plasticity Phenomena
Our analysis of students’ explanations for the four phenotypic plasticity phenomena identified two distinct kinds of mechanistic accounts that broadly framed students’ explanations as: 1) sensing–responding interaction and 2) direct interaction. We next describe in detail these types of accounts and their affordances and constraints for reasoning about phenotypic plasticity phenomena.
Sensing–Responding Accounts
This type of mechanistic account describes the effect of the environment on the phenotype as mediated by a biological sensing–responding mechanism. The environmental stimulus is sensed by a specific biological mechanism, which in turn activates signals that modulate genetic mechanisms. All sensing–responding explanations included a sensing entity and described it as the agency of change that initiated the modulation of the phenotype in response to the environmental stimulus. The environment may change all the time, yet the organism can sense and respond to this change, which results in the modulation and variation of the phenotype. The level at which this modulation occurs varies. Some students suggested a modulation at the organism level (the body shuts off processes), others suggested a mechanism at the cellular level (protein inhibition/activation), and yet others suggested a modulation at the level of gene expression. Some examples for sensing–responding explanations are presented in Table 1.
| Example quote | Analysis |
|---|---|
| “There are sensors. Electrons jump, there is a conformational change that sends a signal and activates a transcription factor that activates genes.” —Yoni, year 2, Tanning | Yoni directly talks about sensors that can sense the environment. He describes how this sensing occurs (electron causes conformational change in a sensing protein) and that it leads to an activation of a response mechanism involving gene expression (activating genes). The sensing mechanisms activate the change, not merely the environment. The modulation is at the level of gene expression. |
| “He doesn’t eat well so it (the body) directs the energy to survival … If there is not enough energy there is a lot of AMP and it can become cAMP and the body understands there is a low amount of energy and tries to store it and use less of it.”—Shay, year 3, Stunting | Shay first suggests that the body “knows” that there is not enough food; thus the body directs what it has to the crucial survival. Based on the comparison between crucial processes that continue and less significant processes that stop, Shay seems to be suggesting that the agency of change is mechanisms that can sense that there is not enough food and react by stopping some processes and not others. In this account, the body is not passive, rather it can sense and respond to the environmental conditions. The response is described at the organism level. |
| “It’s an evolutionary mechanism that is imprinted in the genes for recognizing smells … Receptors and signals. The mother worm ate and got a signal of smell and something links the two (food and smell) … the signal goes to the genetic code and makes it produce proteins for reproducing.”—Mor, year 1, Smell imprinting | Mor is explaining how the sensing of the smell (not only the smell itself) causes the worm to be attracted and to lay more eggs. She describes a sensing mechanism and a signal that alters gene expression. Her description does not include an explanation of how the offspring of the worm are attracted to the smell. When the interviewer draws her attention to that, she says: “I don’t know how … the smell led to a change not in the code but in something that acts on it … how it is read ... but what reads it is also coded … I’m not sure.” Mor could not explain all aspects of the phenomenon, but her explanation is based on a sensing and signaling mechanism by which the organism modulates its response to the environment. |
The sensing–responding account also exemplifies an important characteristic of life—sensing and responding to the environment. Sensing–responding mechanisms are central to core biological concepts such as homeostasis and development (e.g., differentiation). Therefore, a lack of understanding and ability to apply sensing–responding mechanisms is a major impediment to understanding biology.
Direct Interaction Accounts
The second type of mechanistic explanation we identified describes the effect of the environment on phenotypes by focusing on the environment as the agency of change. The organism is not really responding to the environment (or sensing it), but rather the environment acts upon a passive organism. Such explanations describe a chemical or physical interaction between a biological component (organ, tissue, protein, and so on). The environment in these explanations either works in parallel mechanism to the genetic one, while both affect the final phenotype (direct interaction at the phenotype level), directly affects one or more entities in a genetic mechanism and changes them (direct interaction at the cell or protein level), or is enabling the execution of the “genetic plan.” This environmental effect may inhibit or activate entities involved in the molecular and cellular mechanisms leading to the phenotype. Examples for such explanations are presented in Table 2.
| Citation | Analysis |
|---|---|
| “It has to do with the burning of cells. It’s a biological destruction but it has nothing to do with the genetic trait of the skin. Radiation gets in and it ruins a cell, which changes the tissue so it looks darker.”—Mor, year 1, Tanning | Mor is describing a direct interaction at the trait level. The environment (UV radiation) is described as destroying the cells. The environment’s impact is a physical/chemical reaction that would presumably also happen if one exposed dead cells to UV radiation; thus the organism is not actively involved in reacting to the stimulus. The environment essentially acts on a passive organism. |
| “Light is energy, I don’t know maybe it changes a biochemical reaction … the light affects an enzyme … it wakes it up.”—Ela, year 2, Tanning | Ela is describing a direct interaction at the mechanism level. Here, a biological mechanism is activated due to radiation. This is analogous to the effect of temperature on enzymes, the higher the temperature, the faster the enzyme works (until reaching a denaturation point). This effect may also act outside of the in situ context of the cell—the enzyme would also “wake up,” as she phrases it, in a test tube. As in Mor’s explanation, the environment is acting on a seemingly passive organism. It is unclear whether the enzyme that is activated here is part of a responding mechanism. |
| “It’s like in places in which they close girls’ feet in small shoes or put rings on their neck … Even when they are 100 years old their feet are small … so he (a child exhibiting stunted growth) has the genes but doesn’t have the nutrients. His growth plates are trying to grow but do not have nutrients.”—Moti, year 3, Stunting | Moti describes the environment as enabling the execution of the “genetic program.” He compares the effect of malnutrition with a physical environment that does not allow growth. The body itself proceeds with the “genetic plan” of growing, but the environment, namely the lack of nutrients, “stops it.” Again, the environment is acting on a passive organism, and the impact is direct, without building blocks the body cannot grow. |
In all of these examples, the environment, in a sense, “happens” to the organism and modifies it in some way. The organism does not seem to actively sense, respond, or react to the environment.
Figure 1 illustrates the direct interaction mechanism (Figure 1A) in which the environment physically (or chemically) affects one of the entities in the mechanism, and the sensing–responding mechanism (Figure 1B) that involves sensing the environment and responding by modulating mechanisms, leading to phenotypes.
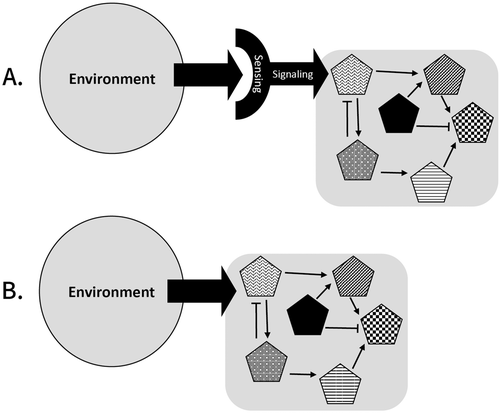
FIGURE 1. Two possible mechanistic accounts for explaining phenotypic plasticity. (A) Sensing–responding mechanism: the environment is sensed via a sensing mechanism that activates a response of modulating a biological mechanism. (B) Direct interaction between the environment and a biological mechanism. Pentagons represent biological entities that are involved in a biological mechanism (gray square).
The four tasks of the interview were all examples of phenotypic plasticity, meaning all involved sensing–responding mechanisms. However, there are cases in which an environmental condition can directly affect the phenotype. For example, hair-straightening chemicals or air humidity level directly modify hair structure. Such cases could be correctly explained by a direct interaction that does not involve any “active” role for the organism. While these are clearly environmental effects on organisms’ phenotypes, these are not a form of phenotypic plasticity, because they do not represent the plasticity of the genotype (and consequently phenotype) but a distinct mechanism that impinged on the biological one.
We next wished to understand the factors that played a role in students’ use of these two types of accounts. We thus further analyzed the association of each account with other factors, such as year of study, biological ideas, and context.
Biological Ideas Associated with Sensing–Responding Explanations
We identified three ideas that were strongly associated with sensing–responding explanations. The first is the idea of proteins as central entities in bimolecular mechanisms. This idea has already been shown to be important for students’ understanding of genetic phenomena (Freidenreich et al., 2011; van Mil et al., 2013; Haskel-Ittah and Yarden, 2017). An analysis of students’ use of this idea in their explanations revealed that many (72%) sensing–responding explanations involved proteins as functional entities in the mechanisms (i.e., entities with relevant activities in the mechanism). Significantly fewer direct interaction explanations (28%, χ2 = 34.69, p < 0.000) involved protein activities, and many of these explanations (32%) did not involve proteins at all.
The second idea we identified is the regulation of gene expression. Regulation of gene expression is one of the response mechanisms triggered by signals from the external/internal environment. Analysis of the association between the idea of regulation of gene expression and the two accounts showed that direct interaction accounts almost never included the idea of gene regulation (only 9% included it). On the contrary, sensing–responding accounts were associated with significantly more descriptions of gene regulation (70% included this idea, χ2 = 66.06, p < 0.0001).
This suggests that the idea of regulation of gene expression was mostly evoked by students when they assumed that the effect of the environment involves sensing–responding mechanisms. This idea is aligned with canonical scientific understandings, and indeed gene regulation is one of the response mechanisms of phenotypic plasticity.
Finally, a third idea that we identified as associated with sensing–responding accounts is the notion of an evolutionary advantage or function. We found this idea in 37 answers (27%), most of which (27 out of 37 answers) were of the sensing–responding type. Here are two representative examples: “UV may cause DNA damage. I would say that this mechanism [tanning as a response to UV light] protects [the organism] from DNA damage” (Larry, third year); “I guess this is a mechanism for survival. Instead of growing, saving the minimum you need for survival” (Noam, second year). Both examples show that, although they were not asked to think of an advantage for the mechanism, some students spontaneously suggested one.
Students Further along in Their Undergraduate Studies Used More Sensing–Responding Explanations
To find out whether students’ level of study (i.e., year in undergraduate studies) influenced their choice of explanation, we analyzed the distribution account types by year of study. Because we presented students with four phenomena and they provided one or two different explanations for each, the scores ranged from 0 to 8 (see Methods).
The distribution of the direct interaction and sensing–responding scores across years of study is presented in Table 3, which shows that third-year students provided significantly more sensing–responding explanations than direct interaction ones (paired t test, p = 0.01, n = 14). First- and second-year students, however, did not seem to significantly privilege one account over the other (paired t test, first year: p = 0.1, n = 9; second year: p = 0.3 n = 15). Table 3 also shows that, while there are significant differences in the average use of sensing–responding accounts across the three years, the differences in the use of direct interaction accounts were not significant (analysis of variance [ANOVA], n = 38, p = 0.03 and p = 1.29).
| Average direct interaction score (SD, number of answers) | Average sensing–responding score (SD, number of answers ) | Paired t test | |
|---|---|---|---|
| Year 1 | 2.33 (1.11, 21) | 1.44 (1.01, 13) | p = 0.1 (T = −1.40) |
| Year 2 | 1.67 (1.04, 25) | 2.27 (1.44, 34) | p = 0.3 (T = 1.04) |
| Year 3 | 1.36 (1.15, 19) | 2.79 (0.80, 39) | p = 0.01 (T = 2.99) |
| ANOVA F (2,35) | p = 1.29 (F = 1.29) | p = 0.03 (F = 3.79) |
As mentioned earlier, all four phenomena presented to the students were of the sensing–responding type. However, students provided different answers to these tasks. To better understand which feature of the questions may have cued students to evoke a certain type of account, we analyzed the distribution of the two accounts across the four cases.
Contextual Features of the Phenomena Affected the Use of the Two Mechanistic Accounts
We expected that differences in contextual features of the phenomena might lead to differences in the prevalence of each type of account. Figure 2 illustrates significant differences between the distributions of the two mechanistic accounts in the four tasks. Some contexts predominantly cued one type of account. By exploring the contextual features of each task, we may explain the distribution of the two accounts for each task.
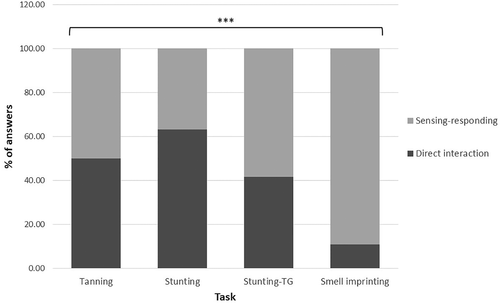
FIGURE 2. Significant differences between the distribution of the two mechanistic accounts in the four cases χ2 = 34.69, p < 0.0001.
The Context of Tanning.
Under this context, the two accounts were equally prevalent. Hence, the transient effect of the environment in humans seemed not to cue a specific mechanistic account.
The Context of Stunting.
The stunting case elicited more direct interaction accounts than sensing–responding accounts. However, adding the information that this stunting phenomenon passed to the next generation (trans-generational stunting) led, in some cases, to a shift from direct interaction to sensing–responding accounts. Overall, 27 students provided a direct interaction account for the stunting task, six of them also suggested a sensing–responding possibility. Five out of these 21 students who provided only a direct interaction account shifted their explanations to a sensing–responding account after being told that this stunting phenomenon passed to the next generation; these five were all third-year students. To better understand why the information about the inheritance of the trait led to a change in students’ answers, we analyzed students’ justifications for changing their answers.
We describe a representative example for a shift from a direct interaction account to a sensing–responding one: Dor, third year, initially said that there are “not enough vitamins and proteins from the food … it is not enough that the genetic potential is there, the right conditions [food] are needed.” After being told of the trans-generational effect, Dor changed his answer to: “It’s a process of atrophy in the genome, an epigenetics process … the child passes it to his offspring … the body does not get nutrients so it can methylate genes that are linked with growth, or ATP use, so in the next generation it already shuts off. Sometimes the methylations are deleted but it probably did not happen.”
For these two third-year students, the context of a trans-generational effect elicited accounts that assumed an impact on genes (because the condition is passed down) and included a sensing–responding mechanism of gene regulation and epigenetic modifications. It might be that, because in trans-generational contexts the environmental stimulus is no longer present, direct interaction accounts appear less likely. The idea of trans-generational effects generates a need for a mechanism that modifies an inherited entity, for example, DNA; therefore, students suggested an epigenetic change in the DNA. Similarly, after being told that stunting is trans-generational, some of the more advanced students also added an epigenetic mechanism to their sensing–responding accounts. This was not the case for the first- and second-year students.
Interestingly, some students did not seem to know that epigenetic changes can pass to future generation, and not all students were familiar with the possibility of epigenetic changes of any sort. Our findings show that in either of these cases, students experienced difficulties in explaining trans-generational plasticity: once students were asked to explain an effect on the next generation, they either said they did not know how to explain it (three students) or suggested a mutation mechanism (four students). Here is a representative example for a shift from a sensing–responding account to an explanation involving mutation (which we classified as direct interaction) that was triggered by adding the information on a trans-generational effect: In the second example, Na’ama, second year, initially claimed that: “If there is not enough food in order to grow and live the body prefers to grow less and to use the energy to survive … there are hormones and signals that are secreted, there is cell communication … the cell stops the process.” Na’ama suggested a regulatory mechanism based on cell communication that stops growth processes as a response to lack of food. After hearing about the trans-generational effect, Na’ama said: “So it is a mutation. Cells are dividing and they don’t have enough substances, so a mutation occurs and this inhibits growth and it persists through generations.” When the interviewer asked whether all people who exhibit stunting have the same mutation, Na’ama replied: “I want to say yes, but the chance for it is very low.”
In these two cases, the inability to explain how a sensing–responding mechanism can persist through generations led to a shift from a sensing–responding explanation to a direct interaction one involving mutations caused directly by the environmental conditions. This direct interaction account was biologically implausible, because it suggested a mutation that specifically affected growth in all individuals under these conditions, meaning a nonrandom mutation. In addition, some students admitted that this targeted mutation idea is incongruent with their prior knowledge; yet they could not think of other possibilities: “It sounds very unrealistic … The DNA is conserved and is not easily changed” (Na’ama, second year, smell imprinting task).
The Context of Smell Imprinting.
Interestingly, the most complex (and likely unfamiliar) phenomenon, smell imprinting, almost exclusively elicited sensing–responding explanations (Figure 2). Two contextual features may explain this result. First, this task deals with a perceptual sense, smelling. It is reasonable to assume that the context of a perceptual smell may cue a sensing feature and thus a sensing–responding explanation. Indeed, half of the students described a receptor that can bind to the smell molecule and transmit a signal that activates certain processes. Hence, it might be that perceptual smell is linked to a molecular sensing entity of a receptor. Second, this task was in the context of a nonhuman organism. This may have led students to suggest mechanisms that they did not think could occur in a human organism. In fact, some students explicitly said that they offered a mechanism that seemed “unreasonable” to them (e.g., the activation of gene-editing mechanisms by the organism itself), but that maybe this could happen in worms.
To summarize, analyzing the differences between the distribution of the two types of mechanistic accounts across the four tasks revealed some of the contextual features that may be affecting students’ answers. More specifically, the organism in the task, the existence of perceptual sense, and the duration of the environmental effect affected the type of account generated (direct interaction or sensing–responding).
DISCUSSION
Two Mechanistic Accounts for Phenotypic Plasticity
In this study, we explored undergraduate students’ explanations for phenotypic plasticity phenomena in which gene–environment interaction leads to a phenotypic change. Phenotypic plasticity involves interactions at different levels between the biological genetically based mechanisms and the environment to modulate the individual’s phenotype.
We identified two types of accounts of phenotypic plasticity. Sensing–responding interaction accounts that involved a mediating biological mechanism that can sense and respond to a stimulus from the environment and direct interaction accounts that involved the environment as directly acting on a passive organism. It seems that, for first- and second-year university students, the direct interaction account is more readily cued, and they tended to invoke it more often. Third-year students provided both accounts, but tended to invoke the sensing–responding account more readily. This may be because they were more familiar with sensing–responding mechanisms from their course work or they were better able to recognize when phenomena involve such mechanisms. Our data do not allow us to distinguish between these two possible explanations. We know that some first- and second-year students were able to generate sensing–responding accounts under some circumstances (trans-generational phenomena); this suggests they were familiar with these types of mechanisms. However, we do not know whether students who did not generate sensing–responding accounts do not know of them (or know them well enough) or whether they simply did not think of them during the interview (or did not think they were appropriate explanations for these phenomena). While the two accounts are both biologically plausible, only the sensing–responding accounts explain phenotypic plasticity. In addition, these sensing–responding capacities of organisms are strongly related to foundational biological concepts such as homeostasis and responding to the environment, key life characteristics.
The importance of the idea of organisms sensing and responding to their environments is not new in biology teaching. In fact, this idea is emphasized in current curricula, in the United States and elsewhere, as part of concepts such as homeostasis, cell communication, and others (National Research Council [NRC], 2012). Yet our findings suggest that even first-year undergraduate students rarely use sensing–responding when explaining interaction with the environment. One of the reasons for this may be that these ideas are presented in the context of body systems and cells as the unit of life (through the concept of homeostasis) and are less frequently presented as a central aspect of genetics instruction (NRC, 2012). Therefore, students may not consider this idea as plausible while reasoning about genetic phenomena.
It should be noted that, without deep domain-specific knowledge, it is not always obvious which kind of explanation is appropriate. For example, the phenomenon of skin wrinkling after prolonged exposure to water (e.g., after swimming) seems like a direct interaction mechanism, much like a pickle is wrinkly in a hypertonic environment because of simple osmosis. Yet the case of wet skin actually results from a reaction of the body’s autonomic nervous system, thus the effect does not occur when there is a nerve damage in the fingers or toes (Lewis, 1935) . This responsive mechanism seemed to have an evolutionary advantage, as it provides a better grip in wet surfaces, much like treads in a car’s tire (Changizi et al., 2011). Thus, without relevant domain-specific knowledge, this example seems like a direct interaction rather a sensing–responding one.
Our results indicate that providing sensing–responding mechanisms at the molecular level was often associated with invoking the idea of proteins as central entities acting at this level. This suggests that knowledge about proteins may be useful for reasoning about mechanisms of phenotypic plasticity. This is in line with growing evidence over the past decade stressing the importance of learning about protein functions in order to support reasoning about a wide variety of genetic phenomena (e.g., Duncan et al., 2011; Todd and Kenyon, 2016; Todd et al., 2019; van Mil et al., 2016; Haskel-Ittah and Yarden, 2017; Haskel-Ittah et al., 2019). Additionally, we found that students also invoked ideas about regulation of gene expression and that such ideas were useful in fleshing out explanations about short- and long-term phenotypic plasticity changes.
In exploring the effect of contextual features of the tasks on the type of account generated, we found that some contextual features in phenomena do cue students (across undergraduate studies) toward a sensing–responding mechanism. For example, the involvement of a perceptual sense (e.g., smelling) seemed to cue more sensing–responding accounts, likely because smelling entails sensing, which is “baked” into the context, thus cuing a sensing–responding mechanism. This may indicate that, when a sensing mechanism is familiar, students identify phenomena as involving sensing–responding more easily.
The fact that the “smelling” case was the only one that involved a nonhuman organism may have also influenced students’ explanations. This may indicate that it is easier for students to suggests unfamiliar sensing–responding mechanisms when the task involves a nonhuman organism.
Interestingly, we found that other contextual features, such as the persistence of phenotypic change, do not always cue students to provide sensing–responding accounts. This means that, unlike experts, students do not always attend to features of the genetic phenomenon that suggest sensing–responding mechanisms; that is, students do not always notice (or understand as such) contextual cues for sensing–responding mechanisms.
Regulation of Gene Expression as Central to Phenotypic Plasticity
An interesting finding of our study is the seeming disconnect between the idea of regulation of gene expression and the phenomenon of phenotypic plasticity, especially for undergraduates in their first years of study. We found that first-year students rarely used the idea of responding to the environment and thus viewed most interaction with the environment as a direct interaction rather than a regulated response. Because gene expression is a means of responding to the environment (external or internal), this raises the question of whether the idea of gene regulation is familiar and meaningful to them. In other words, if they consider most interactions with the environment as happening to the organism rather than the organism responding to the environment, the fact that genes can be turned off or on might not be seen as relevant. One may argue that perhaps they did not learn this idea; however, based on our review of the course syllabi, even first-year students encountered examples of regulation of gene expression in their course work. Moreover, the fact that some of the first- and second-year students who initially provided direct interaction accounts were able to later provide sensing–responding accounts suggests they do understand this idea but did not always draw on it or see it as relevant for the phenomena we provided them.
This finding is important, because regulation of gene expression is often presented in genetics education as an example for how the environment affects genetic traits without emphasizing the sensing and signaling mechanisms that mediate the two (Duncan et al., 2009; NRC, 2012; Boerwinkel et al., 2017). As our results suggest, this link is not obvious or salient to students, and the fact that genes can be turned on or off does not automatically imply (to students) that this switching is a “deliberate” means of response.
Consider Mor’s response, in which she implies that all regulatory mechanisms are themselves proteins that are coded in the genome, and hence she could not understand how the environmental signals can change them (see the third example for sensing–responding explanation, Table 1). This response suggests a conception that genes are both determining traits and regulating themselves without any external signals (no role for the environment).
Conceiving of gene regulation as disconnected from environmental signals is problematic, because it means that the effect of genes on traits is conceived as fixed rather than plastic. This direct connection between genes and traits was indeed shown to strengthen the conception of genes as the ultimate determinant of traits (Parrott and Smith, 2014; Stern and Kampourakis, 2017). One of the dangers of such genetic determinism, aside from it being scientifically incorrect, is that it promotes the conception that variations in behavior, capabilities, or achievements between humans or human groups is genetically based (Lynch et al., 2019)—a conception that was shown to form the basis for racist views and prejudice (Keller, 2005; Dar-Nimrod and Heine, 2011; Donovan, 2016).
While our assumption that conceiving gene regulation as unrelated to sensing and responding to the environment may lead to genetic determinism is speculative, there is some evidence from prior work that lends it some support. For example, there is evidence that knowledge of gene regulation itself is not correlated with deterministic views (Gericke et al., 2017). However, emphasizing the environment’s role in the developmental nature of genetic traits by giving multiple sensing–responding examples, was shown to significantly decrease genetic deterministic views (Jamieson and Radick, 2017). We speculate that the reason for these allegedly contradicting results stem from connecting versus not connecting between gene expression and sensing and responding to the environment.
Implication for Teaching Gene–Environment Interaction
Several implications for genetic instruction and curricula can be drawn from this work. First, it is important to help students understand that interaction with the environment includes both sensing–responding and direct interaction and that phenotypic plasticity includes sensing–responding mechanisms. We claim that understanding under which biological conditions each account is more likely is an important educational goal for biology instruction. This is especially important, given the prevalence and prominence of phenotypic plasticity as opposed to the knowledge students have about this concept (Batzli et al., 2014).
Toward this end, it is important to recognize the effect of context on students’ reasoning about phenotypic plasticity. We advocate providing students with many examples of sensing–responding mechanisms, discussing their general features, and explicitly distinguishing them from direct interaction mechanisms in order to help students develop an understanding of the conditions of applicability that can help them decide when each type of account is appropriate.
Considering our findings, we, in line with others (Duncan et al., 2009; Boerwinkel et al., 2017), are not advocating that students should understand the detailed molecular mechanisms by which the environment affects genetic traits. We nonetheless argue that more emphasis should be placed on teaching sensing mechanisms (including sensing of the internal environment) as part of regulatory mechanisms that are core to genetics. Students should recognize central entities in these mechanisms, such as receptors, and central processes like cellular signaling, regulation of protein activity, and regulation of gene expression.
We also suggest adding discussions of these mechanisms to the genetics curriculum specifically in order to help students understand how the genetic “program” is able to respond to changes in the internal or external environments; in essence, how homeostasis is mediated by mechanisms at the cellular and molecular levels. This emphasis may also assist students in linking genetics to other core concepts in biology.
We also argue that understanding the potential persistence of some environmentally induced epigenetic effects across generations is an important idea, especially from the perspective of social justice, because the inequities in our human-constructed environments impact not only the current generation but may impact those that follow. Moreover, when students do not know of epigenetic mechanisms, or do not see them as relevant, they tend to turn to the idea of mutations. Using mutations to explain phenotypic plasticity phenomena is problematic for two reasons. First, it appears to trigger or bolster alternative conceptions about nonrandom mutations (Albaladejo and Lucas, 1988; Garvin-Doxas and Klymkowsky, 2008). Second, it seems to invoke genetic determinism, as it links permanent changes to the gene with a specific phenotype.
With that said, teaching sensing–responding mechanisms will need to be done carefully in order to avoid entrenching noncanonical views of natural selection. This is because sensing–responding mechanisms might seem somewhat Lamarckian (an intentional trait change for the goal of better fitting the environment). Our finding suggests that students often associated sensing–responding accounts with ideas about evolutionary advantage or a function. We cannot determine whether knowledge about the biological advantage led students to think of a sensing–responding mechanism or if it was the other way around. However, two possibilities may explain the association between the two. First, sensing mechanisms by definition exist in order to better respond and adapt to the changing environment; whereas direct interaction mechanisms can be harmful or beneficial. Thus, thinking about the connection between plasticity and adaptation and an evolutionary advantage may have cued students toward a sensing–responding explanation. Second, sensing and signaling mechanisms have a high energetic cost. In light of evolutionary processes, it makes no senses that such a mechanism survived without providing an evolutionary advantage. Thus, thinking about a sensing–responding mechanism may have cued students to search for a possible evolutionary advantage. This requires consideration of the possibility that a focus on responsiveness at the cellular and genetic levels may reinforce existing misconceptions about natural selection that are difficult to change (Bishop and Anderson, 1990).
To summarize, phenotypic plasticity phenomena seem to cue different explanations that vary across contexts. The mechanisms underlying phenotypic plasticity are likely not automatically constructed by learning about regulatory mechanisms such as regulation of gene expression. That is especially true if only the output of these mechanisms is described (e.g., turning genes on/off) and the sensing and signaling mechanisms that initiated them are ignored. Given the social implications of not understanding the plastic nature of phenotypes, we hope that these findings can help educators and curricular designers develop a more humane genetics curriculum (Donovan et al., 2019).
ACKNOWLEDGMENTS
This work was supported by a Weizmann—Abroad Postdoctoral Grant for Advancing Women in Science.



